Cefiderocol Resistance Conferred by Plasmid-Located Ferric Citrate Transport System in KPC-Producing Klebsiella pneumoniae
- PMID: 39714320
- PMCID: PMC11682805
- DOI: 10.3201/eid3101.241426
Cefiderocol Resistance Conferred by Plasmid-Located Ferric Citrate Transport System in KPC-Producing Klebsiella pneumoniae
Abstract
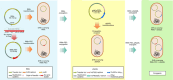
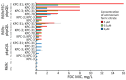
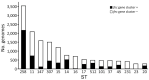
Cefiderocol (FDC), a siderophore-cephalosporin conjugate, is the newest option for treating infection with carbapenem-resistant gram-negative bacteria. We identified a novel mechanism contributing to decreased FDC susceptibility in Klebsiella pneumoniae clinical isolates. The mechanism involves 2 coresident plasmids: pKpQIL, carrying variants of blaKPC carbapenemase gene, and pKPN, carrying the ferric citrate transport (FEC) system. We observed increasing FDC MICs in an Escherichia coli model system carrying different natural pKpQIL plasmids, encoding different K. pneumoniae carbapenemase (KPC) variants, in combination with a conjugative low copy number vector carrying the fec gene cluster from pKPN. We observed transcriptional repression of fiu, cirA, fepA, and fhuA siderophore receptor genes in blaKPC-fec-E. coli cells treated with ferric citrate. Screening of 27,793 K. pneumoniae whole-genome sequences revealed that the fec cluster occurs frequently in some globally distributed different KPC-producing K. pneumoniae clones (sequence types 258, 14, 45, and 512), contributing to reduced FDC susceptibility.
Keywords: Italy; KPC; Klebsiella pneumoniae; antimicrobial resistance; bacteria; carbapenemase-producing Klebsiella pneumoniae; cefiderocol; ferric citrate-dependent iron transport system; siderophore.
Figures




References
-
- World Health Organization. WHO bacterial priority pathogens list, 2024: bacterial pathogens of public health importance to guide research, development and strategies to prevent and control antimicrobial resistance [cited 2024 Nov 27]. https://www.who.int/publications/i/item/9789240093461
-
- Tamma PD, Han JH, Rock C, Harris AD, Lautenbach E, Hsu AJ, et al.; Antibacterial Resistance Leadership Group. Carbapenem therapy is associated with improved survival compared with piperacillin-tazobactam for patients with extended-spectrum β-lactamase bacteremia. Clin Infect Dis. 2015;60:1319–25. 10.1093/cid/civ003 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

