Expanding Polycyclic Tetramate Macrolactam (PoTeM) Core Structure Diversity by Chemo-Enzymatic Synthesis and Bioengineering
- PMID: 39714566
- PMCID: PMC11933527
- DOI: 10.1002/anie.202420335
Expanding Polycyclic Tetramate Macrolactam (PoTeM) Core Structure Diversity by Chemo-Enzymatic Synthesis and Bioengineering
Abstract
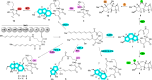
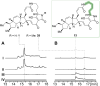
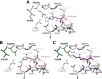
Polycyclic tetramate macrolactams (PoTeMs) represent a growing class of bioactive natural products that are derived from a common tetramate polyene precursor, lysobacterene A, produced by an unusual bacterial iterative polyketide synthase (PKS)/non-ribosomal peptide synthetase (NRPS). The structural and functional diversity of PoTeMs is biosynthetically elaborated from lysobacterene A by pathway-specific cyclizing and modifying enzymes. This results in diverse core structure decoration and cyclization patterns. However, approaches to directly edit the PoTeM carbon skeleton do currently not exist. We thus set out to modify the PoTeM core structure by exchanging the natural l-ornithine-derived building block by l-lysine, hence extending macrocycle size by an additional CH2 group. We developed streamlined synthetic access to lysobacterene A and the corresponding extended analog and achieved cyclization of both precursors by the cognate PoTeM cyclases IkaBC in vitro. This chemo-enzymatic approach corroborated the catalytic competence of IkaBC to produce a larger macrolactam yielding homo-ikarugamycin. We thus engineered the adenylation domain active site of IkaA to directly accept l-lysine, which upon co-expression with IkaBC delivered a recombinant bacterial homo-ikarugamycin producer. Our work establishes an entirely new PoTeM structural framework and sets the stage for the biotechnological diversification of the PoTeM natural product class in general.
Keywords: PoTeM; chemo-enzymatic; natural products; protein engineering; total synthesis.
© 2024 The Author(s). Angewandte Chemie International Edition published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Jomon K., Kuroda Y., Ajisaka M., Sakai H., J. Antibiot. 1972, 25, 271–280. - PubMed
-
- Antosch J., Schaefers F., Gulder T. A. M., Angew. Chem. Int. Ed. 2014, 53, 3011–3014. - PubMed
-
- Zhang G., Zhang W., Zhang Q., Shi T., Ma L., Zhu Y., Li S., Zhang H., Zhao Y.-L., Shi R., Zhang C., Angew. Chem. Int. Ed. 2014, 53, 4840–4844. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

