Machine learning-based identification of histone deacetylase-associated prognostic factors and prognostic modeling for low-grade glioma
- PMID: 39714729
- PMCID: PMC11666852
- DOI: 10.1007/s12672-024-01713-7
Machine learning-based identification of histone deacetylase-associated prognostic factors and prognostic modeling for low-grade glioma
Abstract
Background: Low-grade glioma (LGG) is a slow-growing but invasive tumor that affects brain function. Histone deacetylases (HDACs) play a critical role in gene regulation and tumor progression. This study aims to develop a prognostic model based on HDAC-related genes to aid in risk stratification and predict therapeutic responses.
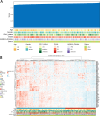
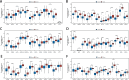
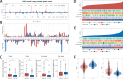
Methods: Expression data from The Cancer Genome Atlas (TCGA) and Chinese Glioma Genome Atlas (CGGA) were analyzed to identify an optimal HDAC-related risk signature from 73 genes using 10 machine learning algorithms. Patients were stratified into high- and low-risk groups based on the median risk score. Prognostic accuracy was evaluated using Kaplan-Meier survival analysis and receiver operating characteristic (ROC) curves. Functional enrichment analyses, including Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), and Gene Set Enrichment Analysis (GSEA), were performed to explore pathways linked to the gene signature. Immune infiltration and tumor microenvironment characteristics were assessed using Single Sample Gene Set Enrichment Analysis (ssGSEA) and ESTIMATE algorithm. SubMap was applied to predict responsiveness to immune checkpoint inhibitors, and chemotherapeutic sensitivity was analyzed via the Genomics of Drug Sensitivity in Cancer (GDSC) database.
Results: A prognostic model consisting of four HDAC-related genes-SP140, BAZ1B, SP100, and SIRT1-was identified. This signature displayed strong prognostic accuracy, achieving a C-index of 0.945. Individuals with LGG were systematically divided into high-risk and low-risk cohorts based on the median risk value, enabling more precise risk stratification. The survival prognosis was significantly worse in the high-risk cohort compared to the low-risk group, highlighting distinct survival trajectories. Notably, the two cohorts exhibited marked shifts in immune checkpoint gene transcriptional profiles and immune cell infiltration maps, underscoring fundamental biological differences that contribute to these differing prognoses.
Conclusion: We developed an HDAC-related four-gene prognostic model that correlates with survival, immune landscape, and therapeutic response in LGG patients. This model may guide personalized treatment strategies and improve prognostic accuracy, warranting further validation in clinical settings.
Keywords: HDAC; Immunotherapy; Low grade glioma; Prognosis; Tumor microenvironment.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests : The authors declare no competing interests.
Figures





References
LinkOut - more resources
Full Text Sources
