Examination of common culture medium for human hepatocytes and engineered heart tissue: Towards an evaluation of cardiotoxicity associated with hepatic drug metabolism in vitro
- PMID: 39715174
- PMCID: PMC11666010
- DOI: 10.1371/journal.pone.0315997
Examination of common culture medium for human hepatocytes and engineered heart tissue: Towards an evaluation of cardiotoxicity associated with hepatic drug metabolism in vitro
Abstract
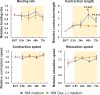
Cardiotoxicity associated with hepatic metabolism and drug-drug interactions is a serious concern. Predicting drug toxicity using animals remains challenging due to species and ethical concerns, necessitating the need to develop alternative approaches. Drug cardiotoxicity associated with hepatic metabolism cannot be detected using a cardiomyocyte-only evaluation system. Therefore, we aimed to establish a system for evaluating cardiotoxicity via hepatic metabolism by co-culturing cryopreserved human hepatocytes (cryoheps) and human iPS cell-derived engineered heart tissues (hiPSC-EHTs) using a stirrer-based microphysiological system. We investigated candidate media to identify a medium that can be used commonly for hepatocytes and cardiomyocytes. We found that the contraction length was significantly greater in the HM Dex (-) medium, the medium used for cryohep culture without dexamethasone, than that in the EHT medium used for hiPSC-EHT culture. Additionally, the beating rate, contraction length, contraction speed, and relaxation speed of hiPSC-EHT cultured in the HM Dex (-) medium were stable throughout the culture period. Among the major CYPs, the expression of CYP3A4 alone was low in cryoheps cultured in the HM Dex (-) medium. However, improved oxygenation using the InnoCell plate increased CYP3A4 expression to levels comparable to those found in the human liver. In addition, CYP3A4 activity was also increased by the improved oxygenation. Furthermore, expression levels of hepatic function-related gene and nuclear receptors in cryoheps cultured in HM Dex (-) medium were comparable to those in the human liver. These results suggest that the HM Dex (-) medium can be applied to co-culture and may allow the evaluation of cardiotoxicity via hepatic metabolism. Moreover, CYP induction by typical inducers was confirmed in cryoheps cultured in the HM Dex (-) medium, suggesting that drug-drug interactions could also be evaluated using this medium. Our findings may facilitate the evaluation of cardiotoxicity via hepatic metabolism, potentially reducing animal testing, lowering costs, and expediting drug development.
Copyright: © 2024 Horiuchi et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures






References
-
- Smyth DH. Alternatives to animal experiments. London: Scolar Press; 1978.
-
- Akingbasote J, Szlapinski S, Hilmas E, Miller P, Rine N. Therapeutic drug monitoring and toxicology: relevance of measuring metabolites. In: Amponsah SK, Pathak YV, editors. Recent advances in therapeutic drug monitoring and clinical toxicology. Cham: Springer; 2022. doi: 10.1007/978-3-031-12398-6_13 - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

