Evaluation and comparison of methods for neuronal parameter optimization using the Neuroptimus software framework
- PMID: 39715260
- PMCID: PMC11706405
- DOI: 10.1371/journal.pcbi.1012039
Evaluation and comparison of methods for neuronal parameter optimization using the Neuroptimus software framework
Abstract
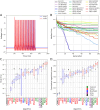
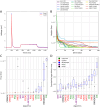
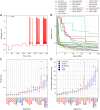
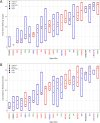
Finding optimal parameters for detailed neuronal models is a ubiquitous challenge in neuroscientific research. In recent years, manual model tuning has been gradually replaced by automated parameter search using a variety of different tools and methods. However, using most of these software tools and choosing the most appropriate algorithm for a given optimization task require substantial technical expertise, which prevents the majority of researchers from using these methods effectively. To address these issues, we developed a generic platform (called Neuroptimus) that allows users to set up neural parameter optimization tasks via a graphical interface, and to solve these tasks using a wide selection of state-of-the-art parameter search methods implemented by five different Python packages. Neuroptimus also offers several features to support more advanced usage, including the ability to run most algorithms in parallel, which allows it to take advantage of high-performance computing architectures. We used the common interface provided by Neuroptimus to conduct a detailed comparison of more than twenty different algorithms (and implementations) on six distinct benchmarks that represent typical scenarios in neuronal parameter search. We quantified the performance of the algorithms in terms of the best solutions found and in terms of convergence speed. We identified several algorithms, including covariance matrix adaptation evolution strategy and particle swarm optimization, that consistently, without any fine-tuning, found good solutions in all of our use cases. By contrast, some other algorithms including all local search methods provided good solutions only for the simplest use cases, and failed completely on more complex problems. We also demonstrate the versatility of Neuroptimus by applying it to an additional use case that involves tuning the parameters of a subcellular model of biochemical pathways. Finally, we created an online database that allows uploading, querying and analyzing the results of optimization runs performed by Neuroptimus, which enables all researchers to update and extend the current benchmarking study. The tools and analysis we provide should aid members of the neuroscience community to apply parameter search methods more effectively in their research.
Copyright: © 2024 Mohácsi et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures




References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

