Predicting the infecting dengue serotype from antibody titre data using machine learning
- PMID: 39715263
- PMCID: PMC11706371
- DOI: 10.1371/journal.pcbi.1012188
Predicting the infecting dengue serotype from antibody titre data using machine learning
Abstract
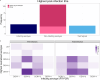
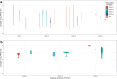
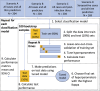
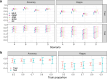
The development of a safe and efficacious vaccine that provides immunity against all four dengue virus serotypes is a priority, and a significant challenge for vaccine development has been defining and measuring serotype-specific outcomes and correlates of protection. The plaque reduction neutralisation test (PRNT) is the gold standard assay for measuring serotype-specific antibodies, but this test cannot differentiate homotypic and heterotypic antibodies and characterising the infection history is challenging. To address this, we present an analysis of pre- and post-infection antibody titres measured using the PRNT, collected from a prospective cohort of Thai children. We applied four machine learning classifiers and multinomial logistic regression to the titre data to predict the infecting serotype. The models were validated against the true infecting serotype, identified using RT-PCR. Model performance was calculated using 100 bootstrap samples of the train and out-of-sample test sets. Our analysis showed that, on average, the greatest change in titre was against the infecting serotype. However, in 53.4% (109/204) of the subjects, the highest titre change did not correspond to the infecting serotype, including in 34.3% (11/35) of dengue-naïve individuals (although 8/11 of these seronegative individuals were seropositive to Japanese encephalitis virus prior to their infection). The highest post-infection titres of seropositive cases were more likely to match the serotype of the highest pre-infection titre than the infecting serotype, consistent with antigenic seniority or cross-reactive boosting of pre-infection titres. Despite these challenges, the best performing machine learning algorithm achieved 76.3% (95% CI 57.9-89.5%) accuracy on the out-of-sample test set in predicting the infecting serotype from PRNT data. Incorporating additional spatiotemporal data improved accuracy to 80.6% (95% CI 63.2-94.7%), while using only post-infection titres as predictor variables yielded an accuracy of 71.7% (95% CI 57.9-84.2%). These results show that machine learning classifiers can be used to overcome challenges in interpreting PRNT titres, making them useful tools in investigating dengue immune dynamics, infection history and identifying serotype-specific correlates of protection, which in turn can support the evaluation of clinical trial endpoints and vaccine development.
Copyright: © 2024 Cracknell Daniels et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
I have read the journal’s policy and the authors of this manuscript have the following competing interests: AF declares travel support from Janssen Pharmaceuticals to attend an industry-sponsored symposium related to dengue. DATC declares Merck and Pfizer contracts, US NIH grants and a US NSF grant to his institution for unrelated work. ID declares consultancy to the WHO and Gavi the vaccine alliance.
Figures

References
-
- Buddhari D, Aldstadt J, Endy TP, Srikiatkhachorn A, Thaisomboonsuk B, Klungthong C, et al.. Dengue virus neutralizing antibody levels associated with protection from infection in Thai cluster studies. PLOS Neglected Tropical Diseases. 2014. Oct 16;8(10):e3230. doi: 10.1371/journal.pntd.0003230 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

