Investigating the Impact of Whole-Genome Duplication on Transposable Element Evolution in Teleost Fishes
- PMID: 39715451
- PMCID: PMC11785729
- DOI: 10.1093/gbe/evae272
Investigating the Impact of Whole-Genome Duplication on Transposable Element Evolution in Teleost Fishes
Abstract
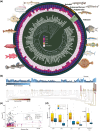
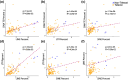
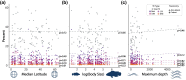
Transposable elements (TEs) can make up more than 50% of any given vertebrate's genome, with substantial variability in TE composition among lineages. TE variation is often linked to changes in gene regulation, genome size, and speciation. However, the role that genome duplication events have played in generating abrupt shifts in the composition of the mobilome over macroevolutionary timescales remains unclear. We investigated the degree to which the teleost genome duplication (TGD) shaped the diversification trajectory of the teleost mobilome. We integrate a new high coverage genome of Polypterus bichir with data from over 100 publicly available actinopterygian genomes to assess the macroevolutionary implications of genome duplication events on TE evolution in teleosts. Our results provide no evidence for a substantial shift in mobilome composition following the TGD event. Instead, the diversity of the teleost mobilome appears to have been shaped by a history of lineage-specific shifts in composition that are not correlated with commonly evoked drivers of diversification such as body size, water column usage, or latitude. Collectively, these results provide additional evidence for an emerging perspective that TGD did not catalyze bursts of diversification and innovation in the actinopterygian mobilome.
Keywords: Actinopterygii; Teleostei; living fossil; teleost genome duplication; transposable elements.
© The Author(s) 2024. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

