Genome-wide CRISPR/Cas9 screen identifies AraC-daunorubicin-etoposide response modulators associated with outcomes in pediatric AML
- PMID: 39715471
- PMCID: PMC11914169
- DOI: 10.1182/bloodadvances.2024014157
Genome-wide CRISPR/Cas9 screen identifies AraC-daunorubicin-etoposide response modulators associated with outcomes in pediatric AML
Abstract
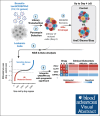
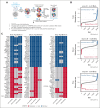
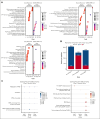
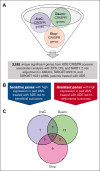
Cytarabine, daunorubicin, and etoposide (ADE) have been the standard backbone of induction chemotherapy regimen for patients with pediatric acute myeloid leukemia (pAML) for >5 decades. However, chemoresistance is still a major concern, and a significant proportion of pAML becomes resistant to ADE treatment and relapse, leading to poor survival. Therefore, there is a considerable need to identify mechanisms mediating drug resistance for overcoming chemoresistance. Herein, we performed synthetic lethal CRISPR/Cas9 screens using the ADE components to identify response markers. We further integrated significant markers in 3 independent pAML clinical cohorts treated with only an ADE regimen to identify drug response biomarkers with prognostic significance. We were able to identify several mediators that represent clinically and biologically significant marker genes for ADE treatment, such as BCL2, CLIP2, and VAV3, which are resistant markers to ADE, with high expression associated with poor outcomes in pAML treated with ADE, and GRPEL1, HCFC1, and TAF10, which are sensitive markers to ADE, with high expression showing beneficial outcomes. Notably, BCL2, CLIP2, and VAV3 knockdowns in their expression in AML cell lines sensitized the cells more to the ADE components, suggesting that these modulators should be further studied as potential therapeutic targets to overcome chemoresistance.
© 2025 American Society of Hematology. Published by Elsevier Inc. Licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0), permitting only noncommercial, nonderivative use with attribution. All other rights reserved.
Conflict of interest statement
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Figures



References
-
- Bose P, Vachhani P, Cortes JE. Treatment of relapsed/refractory acute myeloid leukemia. Curr Treat Options Oncol. 2017;18(3):17. - PubMed
-
- Rubnitz JE, Kaspers GJL. How I treat pediatric acute myeloid leukemia. Blood. 2021;138(12):1009–1018. - PubMed
-
- American Cancer Society Cancer facts & figures 2022. https://www.cancer.org/research/cancer-facts-statistics/all-cancer-facts...
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

