Development of mirror-image monobodies targeting the oncogenic BCR::ABL1 kinase
- PMID: 39715735
- PMCID: PMC11666773
- DOI: 10.1038/s41467-024-54901-y
Development of mirror-image monobodies targeting the oncogenic BCR::ABL1 kinase
Abstract
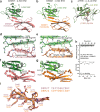
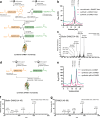
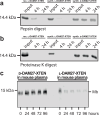
Mirror-image proteins, composed of D-amino acids, are an attractive therapeutic modality, as they exhibit high metabolic stability and lack immunogenicity. Development of mirror-image binding proteins is achieved through chemical synthesis of D-target proteins, phage display library selection of L-binders and chemical synthesis of (mirror-image) D-binders that consequently bind the physiological L-targets. Monobodies are well-established synthetic (L-)binding proteins and their small size (~90 residues) and lack of endogenous cysteine residues make them particularly accessible to chemical synthesis. Here, we develop monobodies with nanomolar binding affinities against the D-SH2 domain of the leukemic tyrosine kinase BCR::ABL1. Two crystal structures of heterochiral monobody-SH2 complexes reveal targeting of the pY binding pocket by an unconventional binding mode. We then prepare potent D-monobodies by either ligating two chemically synthesized D-peptides or by self-assembly without ligation. Their proper folding and stability are determined and high-affinity binding to the L-target is shown. D-monobodies are protease-resistant, show long-term plasma stability, inhibit BCR::ABL1 kinase activity and bind BCR::ABL1 in cell lysates and permeabilized cells. Hence, we demonstrate that functional D-monobodies can be developed readily. Our work represents an important step towards possible future therapeutic use of D-monobodies when combined with emerging methods to enable cytoplasmic delivery of monobodies.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: A.Ko. and S.K. are listed as inventors on issued and pending patents on the monobody technology filed by the University of Chicago (US Patent 9512199 B2 and related pending applications). The other authors declare no competing interests.
Figures






References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Miscellaneous

