Chromosome architecture and low cohesion bias acrocentric chromosomes towards aneuploidy during mammalian meiosis
- PMID: 39715766
- PMCID: PMC11666783
- DOI: 10.1038/s41467-024-54659-3
Chromosome architecture and low cohesion bias acrocentric chromosomes towards aneuploidy during mammalian meiosis
Abstract
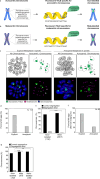
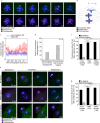
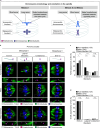
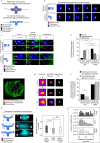
Aneuploidy in eggs is a leading cause of miscarriages or viable developmental syndromes. Aneuploidy rates differ between individual chromosomes. For instance, chromosome 21 frequently missegregates, resulting in Down Syndrome. What causes chromosome-specific aneuploidy in meiosis is unclear. Chromosome 21 belongs to the class of acrocentric chromosomes, whose centromeres are located close to the chromosome end, resulting in one long and one short chromosome arm. We demonstrate that acrocentric chromosomes are generally more often aneuploid than metacentric chromosomes in porcine eggs. Kinetochores of acrocentric chromosomes are often partially covered by the short chromosome arm during meiosis I in human and porcine oocytes and orient less efficiently toward the spindle poles. These partially covered kinetochores are more likely to be incorrectly attached to the spindle. Additionally, sister chromatids of acrocentric chromosomes are held together by lower levels of cohesin, making them more vulnerable to age-dependent cohesin loss. Chromosome architecture and low cohesion therefore bias acrocentric chromosomes toward aneuploidy during mammalian meiosis.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


References
-
- Charalambous, C., Webster, A. & Schuh, M. Aneuploidy in mammalian oocytes and the impact of maternal ageing. Nat. Rev. Mol. Cell Biol. 24, 27–44 (2023). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

