Integrated analysis of cell cycle and p53 signaling pathways related genes in breast, colorectal, lung, and pancreatic cancers: implications for prognosis and drug sensitivity for therapeutic potential
- PMID: 39715832
- PMCID: PMC11666898
- DOI: 10.1007/s12672-024-01712-8
Integrated analysis of cell cycle and p53 signaling pathways related genes in breast, colorectal, lung, and pancreatic cancers: implications for prognosis and drug sensitivity for therapeutic potential
Abstract
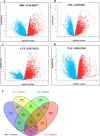
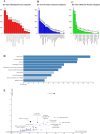
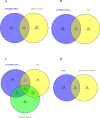
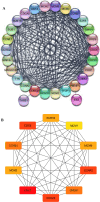
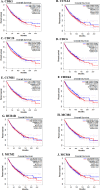
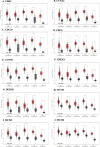
Cancer, a leading cause of death worldwide, is projected to increase by 76.6% in new cases and 89.7% in mortality by 2050 (WHO 2022). Among various types, lung cancer is the most prevalent with high morbidity, while breast, colorectal, and pancreatic cancers also show high mortality rates. Cancer progression often involves disruption in cell cycle regulation and signaling pathways, with mutations in genes like TP53, EGFR, and K-RAS playing significant roles. In this study, we analyzed gene expression datasets to identify common molecular signatures across breast, colorectal, lung and pancreatic cancers. Our focus was on genes related to cell cycle regulation and p53 signaling pathway, intending to discover potential biomarkers for improved diagnosis and treatment strategies. The study analyzed GEO datasets; GSE45827, GSE9348, GSE30219, and GSE62165 for breast, colorectal, lung, and pancreatic cancers respectively. Differentially expressed genes (DEGs) were identified using GEO2R, and functional annotation and pathway analysis were performed using WebGestalt. Common cell cycle and p53 signaling genes were acquired from MSigDB using GSEA. A protein-protein interaction network was constructed using STRING and Cytoscape, identifying top hub genes. Validation of hub genes at mRNA and protein levels was done via GEPIA2 and Human Protein Atlas. Survival analysis was conducted using TCGA data by GEPIA2 and LASSO, and drug sensitivity was analyzed with the GSCA drug bank database, highlighting potential therapeutic targets. The study identified 411 common DEGs among these four cancers. Pathway and functional enrichment revealed key biological processes and pathways like p53 signaling, and cell cycle. The intersection of these DEGs with genes involved in cell cycle and p53 signaling, identified 23 common genes that were used for constructing a PPI network. The top 10 hub genes were validated both for mRNA and protein expression, revealing they are significantly overexpressed in all studied cancers. Prognostic relevance showed that MCM4, MCM6, CCNA2, CDC20, and CHEK1 are associated with survival. Additionally, drug sensitivity analysis highlighted key gene-drug interactions, suggesting potential targets for therapeutic intervention.
Keywords: Breast cancer; Cell cycle; Colorectal cancer; Lung cancer; Pancreatic cancer; p53 signaling.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: In this study, we utilized publicly available datasets. So ethical approval is not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures







References
-
- World Cancer Day 2024: Close the care gap - PAHO/WHO | Pan American Health Organization. https://www.paho.org/en/campaigns/world-cancer-day-2024-close-care-gap.
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
