ADNP is essential for sex-dependent hippocampal neurogenesis, through male unfolded protein response and female mitochondrial gene regulation
- PMID: 39715923
- PMCID: PMC12092271
- DOI: 10.1038/s41380-024-02879-w
ADNP is essential for sex-dependent hippocampal neurogenesis, through male unfolded protein response and female mitochondrial gene regulation
Abstract
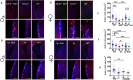
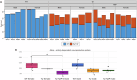
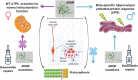
Essential for brain formation and protective against tauopathy, activity-dependent neuroprotective protein (ADNP) is critical for neurogenesis and cognitive functions, while regulating steroid hormone biogenesis. As such, de novo mutations in ADNP lead to syndromic autism and somatic ADNP mutations parallel Alzheimer's disease progression. Furthermore, clinical trials with the ADNP fragment NAP (the investigational drug davunetide) showed efficacy in women suffering from the tauopathy progressive supranuclear palsy and differentially boosted memory in men (spatial) and women (verbal), exhibiting prodromal Alzheimer's disease. While autism is more prevalent in boys and Alzheimer's disease in women, both involve impaired neurogenesis. Here, we asked whether ADNP sex-dependently regulates neurogenesis. Using bromodeoxyuridine (BrdU) as a marker of neurogenesis, we identified two-fold higher labeling in the hippocampal sub-ventricular zone of ADNP-intact male versus female mice. Adnp haplo-insufficient (Adnp+/-) mice or mice CRSIPR/Cas9-edited to present the most prevalent neurodevelopmental ADNP syndrome mutation, p.Tyr718* (Tyr) showed dramatic reductions in male BrdU incorporation, resulting in mutated females presenting higher labeling than males. Treatment with NAP compensated for the male reduction of BrdU labeling. Mechanistically, hippocampal RNAseq revealed male-specific Tyr down-regulation of endoplasmic reticulum unfolded protein response genes critical for sex-dependent organogenesis. Newly discovered mitochondrial accessibility of ADNP was inhibited by the Tyr718* mutation further revealing female-specific Tyr downregulation of mitochondrial ATP6. NAP moderated much of the differential expression caused by p.Tyr718*, accompanied by the down-regulation of neurotoxic, pro-inflammatory and pro-apoptotic genes. Thus, ADNP is a key regulator of sex-dependent neurogenesis that acts by controlling canonical pathways, with NAP compensating for fundamental ADNP deficiencies, striding toward clinical development targeting the ADNP syndrome and related neurodevelopmental/neurodegenerative diseases.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The use of davunetide is under patent protection. Professor Illana Gozes serves as Vice President Drug Development, Exonavis Therapeutics Ltd. Ethical approval & consent to approval: Animal procedures were approved by the institutional animal care and use committee of Tel Aviv University and the Israeli Ministry of Health. All methods were performed in accordance with the relevant guidelines and regulations. No human studies were performed (not applicable).
Figures



Similar articles
-
Activity-dependent neuroprotective protein (ADNP)-end-binding protein (EB) interactions regulate microtubule dynamics toward protection against tauopathy.Prog Mol Biol Transl Sci. 2021;177:65-90. doi: 10.1016/bs.pmbts.2020.07.008. Epub 2020 Aug 14. Prog Mol Biol Transl Sci. 2021. PMID: 33453943 Review.
-
Novel ADNP Syndrome Mice Reveal Dramatic Sex-Specific Peripheral Gene Expression With Brain Synaptic and Tau Pathologies.Biol Psychiatry. 2022 Jul 1;92(1):81-95. doi: 10.1016/j.biopsych.2021.09.018. Epub 2021 Sep 28. Biol Psychiatry. 2022. PMID: 34865853
-
Activity-dependent neuroprotective protein (ADNP) exhibits striking sexual dichotomy impacting on autistic and Alzheimer's pathologies.Transl Psychiatry. 2015 Feb 3;5(2):e501. doi: 10.1038/tp.2014.138. Transl Psychiatry. 2015. PMID: 25646590 Free PMC article.
-
Discovery of autism/intellectual disability somatic mutations in Alzheimer's brains: mutated ADNP cytoskeletal impairments and repair as a case study.Mol Psychiatry. 2021 May;26(5):1619-1633. doi: 10.1038/s41380-019-0563-5. Epub 2019 Oct 30. Mol Psychiatry. 2021. PMID: 31664177 Free PMC article.
-
Sexual divergence in activity-dependent neuroprotective protein impacting autism, schizophrenia, and Alzheimer's disease.J Neurosci Res. 2017 Jan 2;95(1-2):652-660. doi: 10.1002/jnr.23808. J Neurosci Res. 2017. PMID: 27870441 Review.
Cited by
-
Editorial: A Child with ADNP Syndrome: A Case Study of Symptoms, Diagnostic Process and Innovative Behavioral Intervention Modes.J Mol Neurosci. 2025 Apr 24;75(2):53. doi: 10.1007/s12031-025-02344-5. J Mol Neurosci. 2025. PMID: 40272624 No abstract available.
References
-
- Mandel S, Rechavi G, Gozes I. Activity-dependent neuroprotective protein (ADNP) differentially interacts with chromatin to regulate genes essential for embryogenesis. Dev Biol. 2007;303:814–24. - PubMed
-
- Bassan M, Zamostiano R, Davidson A, Pinhasov A, Giladi E, Perl O, et al. Complete sequence of a novel protein containing a femtomolar-activity-dependent neuroprotective peptide. J Neurochem. 1999;72:1283–93. - PubMed
-
- Mandel S, Gozes I. Activity-dependent neuroprotective protein constitutes a novel element in the SWI/SNF chromatin remodeling complex. J Biol Chem. 2007;282:34448–56. - PubMed
-
- Zamostiano R, Pinhasov A, Gelber E, Steingart RA, Seroussi E, Giladi E, et al. Cloning and characterization of the human activity-dependent neuroprotective protein. J Biol Chem. 2001;276:708–14. - PubMed
-
- Oz S, Kapitansky O, Ivashco-Pachima Y, Malishkevich A, Giladi E, Skalka N, et al. The NAP motif of activity-dependent neuroprotective protein (ADNP) regulates dendritic spines through microtubule end binding proteins. Mol psychiatry. 2014;19:1115–24. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

