Fermentation process of tobacco leaves drives the specific changes of microbial community
- PMID: 39716094
- PMCID: PMC11664857
- DOI: 10.1186/s12866-024-03702-w
Fermentation process of tobacco leaves drives the specific changes of microbial community
Abstract
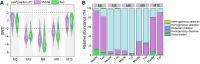
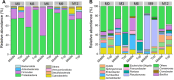
Background: The changes of microbial community on tobacco leaves are affected by several factors during fermentation. However, the relative contribution of different factors in determining microbial community is not clear. This study investigated the effects of fermentation time (fermentation for 0, 3, 6, 9 and 12 months), leaf position (middle and top tobacco leaves) and fermentation site (Longyan and Xiamen warehouses) on bacterial community of tobacco leaves using 16 S rDNA sequencing.
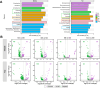
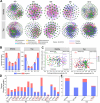
Results: The results demonstrated that fermentation time had a much stronger impact on bacterial diversity, composition, co-occurrence network and functional profiles than leaf position and fermentation site. With the fermentation progressed, the difference of bacterial community between middle and top tobacco leaves was gradually reduced or even disappeared. The bacterial community diversity and network complexity at three, six and nine months of fermentation were significantly lower than those at fermentation initiation. Specific bacterial genera with desired functions were recruited at different fermentation stages, such as Terribacillus, Pantoea and Franconibacter at three or six months of fermentation and Pseudomonas at nine months of fermentation. The recruited microorganisms would form biofilms on tobacco leaves and compete for polysaccharide or protein substances to accelerate the degradation of tobacco macromolecular substances.
Conclusions: In conclusion, fermentation time was an important factor in determining the composition and function of microbial community on tobacco leaves during the fermentation process.
Keywords: Co-occurrence network; Functional profiles; Microbial community; Plant fermentation; Tobacco leaves.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures


References
-
- Chen SS, Fu Y, Bian XQ, Zhao M, Zuo YL, Ge YH, et al. Investigation and dynamic profiling of oligopeptides, free amino acids and derivatives during pu-erh tea fermentation by ultra-high performance liquid chromatography tandem mass spectrometry. Food Chem. 2022;371:371. 10.1016/j.foodchem.2021.131176. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

