The expression of pro-prion, a transmembrane isoform of the prion protein, leads to the constitutive activation of the canonical Wnt/β-catenin pathway to sustain the stem-like phenotype of human glioblastoma cells
- PMID: 39716276
- PMCID: PMC11667964
- DOI: 10.1186/s12935-024-03581-1
The expression of pro-prion, a transmembrane isoform of the prion protein, leads to the constitutive activation of the canonical Wnt/β-catenin pathway to sustain the stem-like phenotype of human glioblastoma cells
Abstract
Background: Cellular prion protein (PrPC) is a widely expressed membrane-anchored glycoprotein, which has been associated with the development and progression of several types of human malignancies, controlling cancer stem cell activity. However, the different molecular mechanisms regulated by PrPC in normal and tumor cells have not been characterized yet.
Methods: To assess the role of PrPC in patient-derived glioblastoma stem cell (GSC)-enriched cultures, we generated cell lines in which PrPC was either overexpressed or down-regulated and investigated, in 2D and 3D cultures, its role in cell proliferation, migration, and invasion. We evaluated the role of PrPC in supporting GSC stemness and the intracellular signaling involved using qRT-PCR, immunocytofluorescence, and Western blot.
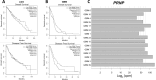
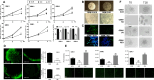
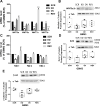
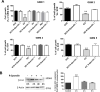
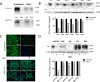
Results: Stable PrPC down-regulation leads to a significant reduction of GSC proliferation, migration, and invasiveness. These effects were associated with the inhibition of the expression of stemness genes and overexpression of differentiation markers. At molecular level PrPC down-regulation caused a significant inhibition of Wnt/β-catenin pathway, through a reduced expression of Wnt and Frizzled ligand/receptor subtypes, resulting in the inhibition of β-catenin transcriptional activity, as demonstrated by the reduced expression of its target genes. The specificity of PrPC in these effects was demonstrated by rescuing the phenotype and the biological activity of PrPC down-regulated GSCs by re-expressing the protein. To get insights into the distinct mechanisms by which PrPC regulates proliferation in GSCs, but not in normal astrocytes, we analyzed structural features of PrPC in glioma stem cells and astrocytes using Western blot and immunofluorescence techniques. Using Pi-PLC, an enzyme that cleaves GPI anchors, we show that, in GSCs, PrP is retained within the plasma membrane in an immature Pro-PrP isoform whereas in astrocytes, it is expressed in its mature PrPC form, anchored on the extracellular face of the plasma membrane.
Conclusions: The persistence of Pro-PrP in GSCs is an altered cellular mechanism responsible of the aberrant, constitutive activation of Wnt/β-catenin pathway, which contributes to glioblastoma malignant features. Thus, the activity of Pro-PrP may represent a targetable vulnerability in glioblastoma cells, offering a novel approach for differentiating and eradicating glioblastoma stem cells.
Keywords: Glioblastoma stem cells; Prion protein; Pro-prion, Wnt/β-catenin pathway..
© 2024. The Author(s).
Conflict of interest statement
Declarations. Ethics Declaration: Human GBM specimens were obtained from the Neurosurgery Dept. of IRCCS Ospedale Policlinico San Martino (Genova, Italy), after Institutional Ethical Committee approval (CER Liguria 360/2019) and patients’ informed consent. Competing interests: The authors declare no competing interests.
Figures


References
Grants and funding
- (DN. 1553 11.10.2022)/#NEXTGENERATIONEU (NGEU) and funded by the Ministry of University and Research (MUR), National Recovery and Resilience Plan (NRRP), project MNESYS (PE0000006) - A Multiscale integrated approach to the study of the nervous system in health and disease (DN. 1553 11.10.2022)
- (grant 2018)/"Fondazione Giovanni Celeghin Contro i Tumori Cerebrali"
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

