Comparative lipidomic and metabolomic profiling of mdx and severe mdx-apolipoprotein e-null mice
- PMID: 39716324
- PMCID: PMC11664822
- DOI: 10.1186/s13395-024-00368-w
Comparative lipidomic and metabolomic profiling of mdx and severe mdx-apolipoprotein e-null mice
Abstract
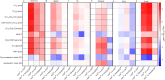
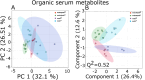
Despite its notoriously mild phenotype, the dystrophin-deficient mdx mouse is the most common model of Duchenne muscular dystrophy (DMD). By mimicking a human DMD-associated metabolic comorbidity, hyperlipidemia, in mdx mice by inactivating the apolipoprotein E gene (mdx-ApoE) we previously reported severe myofiber damage exacerbation via histology with large fibro-fatty infiltrates and phenotype humanization with ambulation dysfunction when fed a cholesterol- and triglyceride-rich Western diet (mdx-ApoEW). Herein, we performed comparative lipidomic and metabolomic analyses of muscle, liver and serum samples from mdx and mdx-ApoEW mice using solution and high-resolution-magic angle spinning (HR-MAS) 1H-NMR spectroscopy. Compared to mdx and regular chow-fed mdx-ApoE mice, we observed an order of magnitude increase in lipid deposition in gastrocnemius muscle of mdx-ApoEW mice including 11-fold elevations in -CH3 and -CH2 lipids, along with pronounced elevations in serum cholesterol, fatty acid, triglyceride and phospholipids. Hepatic lipids were also elevated but did not correlate with the extent of muscle lipid infiltration or differences in serum lipids. This study provides the first lipometabolomic signature of severe mdx lesions exacerbated by high circulating lipids and lends credence to claims that the liver, the main regulator of whole-body lipoprotein metabolism, may play only a minor role in this process.
Keywords: Duchenne; Fibrofatty infiltration; HR-MAS; Liver; Metabolism; Muscle; NMR.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: This study was approved by the University of Florida (Gainesville, FL) and University of British Columbia Institutional Animal Care and Use Committees. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures




References
-
- Sicinski P, Geng Y, Ryder-Cook AS, Barnard EA, Darlison MG. Barnard P.J. The molecular basis of muscular dystrophy in the (mdx) mouse. A point mutation. Science. 1989;244:1578–80. - PubMed
-
- Townsend D, Yasuda S, Metzger J. Cardiomyopathy of Duchenne muscular dystrophy: pathogenesis and prospect of membrane sealants as a new therapeutic approach. Expert Rev Cardiovasc Ther. 2007;5(1):99–109. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

