A genome-scale metabolic reconstruction provides insight into the metabolism of the thermophilic bacterium Rhodothermus marinus
- PMID: 39716382
- PMCID: PMC11730185
- DOI: 10.1093/femsec/fiae167
A genome-scale metabolic reconstruction provides insight into the metabolism of the thermophilic bacterium Rhodothermus marinus
Abstract
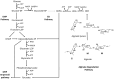
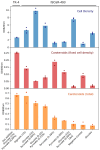
The thermophilic bacterium Rhodothermus marinus has mainly been studied for its thermostable enzymes. More recently, the potential of using the species as a cell factory and in biorefinery platforms has been explored, due to the elevated growth temperature, native production of compounds such as carotenoids and exopolysaccharides, the ability to grow on a wide range of carbon sources including polysaccharides, and available genetic tools. A comprehensive understanding of the metabolism of cell factories is important. Here, we report a genome-scale metabolic model of R. marinus DSM 4252T. Moreover, the genome of the genetically amenable R. marinus ISCaR-493 was sequenced and the analysis of the core genome indicated that the model could be used for both strains. Bioreactor growth data were obtained, used for constraining the model and the predicted and experimental growth rates were compared. The model correctly predicted the growth rates of both strains. During the reconstruction process, different aspects of the R. marinus metabolism were reviewed and subsequently, both cell densities and carotenoid production were investigated for strain ISCaR-493 under different growth conditions. Additionally, the dxs gene, which was not found in the R. marinus genomes, from Thermus thermophilus was cloned on a shuttle vector into strain ISCaR-493 resulting in a higher yield of carotenoids.
Keywords: Rhodothermus marinus; carotenoids; genome-scale metabolic model.
© The Author(s) 2024. Published by Oxford University Press on behalf of FEMS.
Conflict of interest statement
None declared.
Figures

References
-
- Alfredsson GA, Kristjansson JK, Hjrleifsdottir S et al. Rhodothermus marinus, gen.nov., sp. nov., a thermophilic, halophilic bacterium from submarine hot springs in Iceland. J Gen Microbiol. 1988;134:299–306.
-
- Allahgholi L, Sardari RRR, Hakvåg S et al. Composition analysis and minimal treatments to solubilize polysaccharides from the brown seaweed Laminaria digitata for microbial growth of thermophiles. J Appl Phycol. 2020;32:1933–47. 10.1007/s10811-020-02103-6. - DOI
-
- Andrews S. FastQC, Babraham Bioinformatics. 2010. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (1 June 2020, date last accessed).
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

