ID3 promotes erythroid differentiation and is repressed by a TAL1-PRMT6 complex
- PMID: 39716491
- PMCID: PMC11847539
- DOI: 10.1016/j.jbc.2024.108119
ID3 promotes erythroid differentiation and is repressed by a TAL1-PRMT6 complex
Abstract
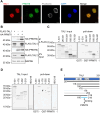
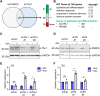
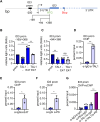
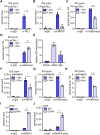
Erythropoiesis is controlled by transcription factors that recruit epigenetic cofactors to establish and maintain erythrocyte-specific gene expression patterns while repressing alternative lineage commitment. The transcription factor TAL1 (T-cell acute lymphocytic leukemia 1) is critical for establishing erythroid gene expression. It acts as an activator or repressor of genes, depending on associated epigenetic cofactors. Understanding the epigenetic function of TAL1 during erythropoiesis is key to improving in vitro erythroid differentiation and understanding pathological erythropoiesis. Therefore, the regulatory mechanisms that control the function of TAL1 during erythropoiesis are under intense investigation. Here, we show that TAL1 interacts with protein-arginine-methyltransferase-6 (PRMT6) on the ID3 (inhibitor-of-DNA-binding-3) gene in K562 and hCD34+ cells. The ID protein family is a critical transcriptional regulator of hematopoietic cell differentiation. We show that TAL1 and PRMT6 are present at the ID3 promoter, and that TAL1 is involved in the recruitment of PRMT6. Here, PRMT6 epigenetically regulates ID3 expression by mediating dimethylation of histone 3 at arginine 2. Thus, TAL1-PRMT6 epigenetically represses ID3 expression in progenitors, which is relieved upon erythroid differentiation, leading to increased expression. Overexpression of ID3 in primary hCD34+ cells enhances erythropoiesis. Our results show that a TAL1-PRMT6 complex regulates genes important for erythropoiesis, such as ID3. Manipulation of ID3 expression may be a way to promote in vitro differentiation of hCD34+ cells into erythrocytes.
Keywords: basic helix–loop–helix transcription factor; cell differentiation; epigenetics; erythropoiesis; histone methylation; transcription corepressor; transcription regulation.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures
References
-
- Orkin S.H. Diversification of haematopoietic stem cells to specific lineages. Nat. Rev. Genet. 2000;1:57–64. - PubMed
-
- Cantor A.B., Orkin S.H. Transcriptional regulation of erythropoiesis: an affair involving multiple partners. Oncogene. 2002;21:3368–3376. - PubMed
-
- Cantor A.B., Orkin S.H. Hematopoietic development: a balancing act. Curr. Opin. Genet. Dev. 2001;11:513–519. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

