Analysis of eIF4E-family members in fungi contributes to their classification in eukaryotes
- PMID: 39716494
- PMCID: PMC11791286
- DOI: 10.1016/j.jbc.2024.108129
Analysis of eIF4E-family members in fungi contributes to their classification in eukaryotes
Abstract
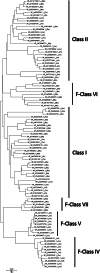
The kingdom of fungi contains highly diverse species. However, fundamental processes sustaining life such as RNA metabolism are much less comparatively studied in Fungi than in other kingdoms. A key factor in the regulation of mRNA expression is the cap-binding protein eIF4E, which plays roles in mRNA nuclear export, storage, and translation. The advent of massive genomics has unveiled a constellation of eIF4E-family members across eukaryotes. However, how this protein diverged into fungal species remains largely unexplored. Here, we studied the genome of 538 species from six evolutionarily distant phyla and retrieved 1462 eIF4E cognates. The analyzed species contained 1 to 7 paralogs. We sorted all cognates in six phylogenetically coherent clades, that we termed Class I to VII (mammalian Class III was absent in Fungi). Proteins from Classes IV to VII did not match the current eIF4Es classification that is based on variations in the residues equivalent to W43 and W56 of the human protein. eIF4Es from other eukaryotes do not fit into this classification either. Thus, we have updated the eIF4E categorization based on cladistics and the presence of cap-binding amino acids to better fit eIF4E's diversity across eukaryotes. Finally, we predicted the structure of the global protein and the cap-binding pocket and experimentally tested the ability to rescue the lack of endogenous eIF4E in Saccharomyces cerevisiae of representative members of each of the six classes of fungal eIF4E.
Keywords: RNA metabolism; eIF4E; fungi; gene expression; mRNA; translation initiation.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures

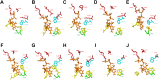

Similar articles
-
eIF4E and Interactors from Unicellular Eukaryotes.Int J Mol Sci. 2020 Mar 21;21(6):2170. doi: 10.3390/ijms21062170. Int J Mol Sci. 2020. PMID: 32245232 Free PMC article. Review.
-
Structural motifs in eIF4G and 4E-BPs modulate their binding to eIF4E to regulate translation initiation in yeast.Nucleic Acids Res. 2018 Jul 27;46(13):6893-6908. doi: 10.1093/nar/gky542. Nucleic Acids Res. 2018. PMID: 30053226 Free PMC article.
-
Phylogenetic analysis of eIF4E-family members.BMC Evol Biol. 2005 Sep 28;5:48. doi: 10.1186/1471-2148-5-48. BMC Evol Biol. 2005. PMID: 16191198 Free PMC article.
-
The alveolate translation initiation factor 4E family reveals a custom toolkit for translational control in core dinoflagellates.BMC Evol Biol. 2015 Feb 10;15(1):14. doi: 10.1186/s12862-015-0301-9. BMC Evol Biol. 2015. PMID: 25886308 Free PMC article.
-
eIF4E as a molecular wildcard in metazoans RNA metabolism.Biol Rev Camb Philos Soc. 2023 Dec;98(6):2284-2306. doi: 10.1111/brv.13005. Epub 2023 Aug 8. Biol Rev Camb Philos Soc. 2023. PMID: 37553111 Review.
References
-
- Hernández G., Vazquez-Pianzola P. eIF4E as a molecular wildcard in metazoans RNA metabolism. Biol. Rev. Cambr. Phil. Soc. 2023;98:2284–2306. - PubMed
-
- Marcotrigiano J., Gingras A.C., Sonenberg N., Burley S.K. Cocrystal structure of the messenger RNA 5' cap-binding protein (eIF4E) bound to 7-methyl-GDP. Cell. 1997;89:951–961. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

