Microglia-Derived Vitamin D Binding Protein Mediates Synaptic Damage and Induces Depression by Binding to the Neuronal Receptor Megalin
- PMID: 39716879
- PMCID: PMC11809382
- DOI: 10.1002/advs.202410273
Microglia-Derived Vitamin D Binding Protein Mediates Synaptic Damage and Induces Depression by Binding to the Neuronal Receptor Megalin
Abstract
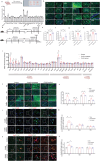
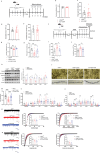
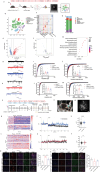
Vitamin D binding protein (VDBP) is a potential biomarker of major depressive disorder (MDD). This study demonstrates for the first time that VDBP is highly expressed in core emotion-related brain regions of mice susceptible to chronic unpredictable mild stress (CUMS). Specifically, the overexpression of microglia (MG)-derived VDBP in the prelimbic leads to depression-like behavior and aggravates CUMS-induced depressive phenotypes in mice, whereas conditional knockout of MG-derived VDBP can reverse both neuronal damage and depression-like behaviors. Mechanistically, the binding of MG-derived VDBP with the neuronal receptor megalin mediates the downstream SRC signaling pathway, leading to neuronal and synaptic damage and depression-like behaviors. These events may be caused by biased activation of inhibitory neurons and excitatory-inhibitory imbalance. Importantly, this study has effectively identified MG-derived VDBP as a pivotal mediator in the interplay between microglia and neurons via its interaction with the neuronal receptor megalin and intricate downstream impacts on neuronal functions, thus offering a promising therapeutic target for MDD.
Keywords: depression; megalin; microglia; neuron damage; vitamin D binding protein.
© 2024 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Shi Y., Song R., Wang L., Qi Y., Zhang H., Zhu J., Zhang X., Tang X., Zhan Q., Zhao Y., Swaab D. F., Bao A.‐M., Zhang Z., J. Affective Disord. 2020, 277, 620. - PubMed
MeSH terms
Substances
Grants and funding
- 2022ZD0211701/China Science and Technology Innovation 2030 - Major Project
- 2021ZD0200700/China Science and Technology Innovation 2030 - Major Project
- 82130042/National Natural Science Foundation of China
- 81830040/National Natural Science Foundation of China
- 82371532/National Natural Science Foundation of China
- GJHZ20210705141400002/Shenzhen Science and Technology Serial Funds
- KCXFZ20211020164543006/Shenzhen Science and Technology Serial Funds
- JCYJ20220818101615033/Shenzhen Science and Technology Serial Funds
- ZDSYS20220606100606014/Shenzhen Science and Technology Serial Funds
- KQTD20221101093608028/Shenzhen Science and Technology Serial Funds
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
