FGF19-Activated Hepatic Stellate Cells Release ANGPTL4 that Promotes Colorectal Cancer Liver Metastasis
- PMID: 39716892
- PMCID: PMC11831508
- DOI: 10.1002/advs.202413525
FGF19-Activated Hepatic Stellate Cells Release ANGPTL4 that Promotes Colorectal Cancer Liver Metastasis
Abstract
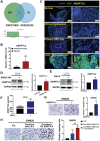
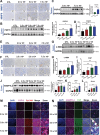
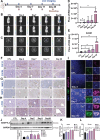
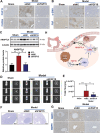
Liver and lung are the most common metastatic sites in colorectal cancer (CRC), where the tumor microenvironment (TME) plays a crucial role in the progression and metastasis of CRC. Understanding the interactions between various types of cells in the TME can suggest innovative therapeutic strategies. Using single-cell RNA sequencing (scRNA-Seq) and clinical samples, fibroblast growth factor-19 (FGF19, rodent FGF15) is found to mediate a significant interaction between CRC cells and cancer-associated fibroblasts (CAFs), activating the hepatic stellate cells (HSCs)-to-CAFs differentiation. In various CRC metastatic mouse models, it is shown that FGF15 has a more pronounced effect on liver metastasis compared to pulmonary metastasis. More importantly, the differentially expressed genes (DEGs) are also identified from the RNA-Seq dataset upon the activation of HSCs by FGF19 and compared the DEGs in matched primary and metastatic mRNA samples from patients with CRC liver metastasis (CRCLM), it is found that the ANGPTL4 gene is significantly associated with HSCs activation. Different mouse models also demonstrated the impact of the FGF19/ANGPTL4 axis on the severity of CRCLM. Importantly, disruption of this axis significantly inhibits CRCLM in vivo. This study is among the first to demonstrate the impact of the FGF19/ANGPTL4 axis on CRCLM, offering a novel therapeutic strategy.
Keywords: ANGPTL4; FGF19; cancer‐associated fibroblasts; colorectal cancer liver metastasis; tumor microenvironment.
© 2024 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
MeSH terms
Substances
Grants and funding
- 2023YFC3502800/National Key Research and Development Program of China
- 82074019/National Natural Science Foundation of China
- 82274158/National Natural Science Foundation of China
- 20222042/Administration of Traditional Chinese Medicine of Guangdong Province
- 20241074/Administration of Traditional Chinese Medicine of Guangdong Province
- 2023KTSCX023/Characteristic Innovation Projects of Universities in Guangdong Province
- 2023A1515011811/Natural Science Foundation of Guangdong Province
- 08193596/Health and Medical Research Fund
- RC-FNRA-IG/23-24/SCM/01/Initial Grant for Faculty Niche Research Areas
- RC-SFCRG/23-24/SCM/02/Seed Funding for Collaborative Research Grants
LinkOut - more resources
Full Text Sources
Medical
