Microphysiological System-Generated Physiological Shear Forces Reduce TNF-α-Mediated Cartilage Damage in a 3D Model of Arthritis
- PMID: 39716911
- PMCID: PMC11831510
- DOI: 10.1002/advs.202412010
Microphysiological System-Generated Physiological Shear Forces Reduce TNF-α-Mediated Cartilage Damage in a 3D Model of Arthritis
Abstract
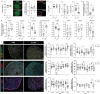
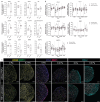
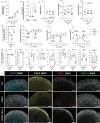
Osteoarthritis (OA) is a leading cause of disability, often resulting from overuse or injury, but inactivity can also contribute to cartilage degeneration. Conventional in vivo models struggle to isolate and study the specific effects of mechanical stress on cartilage health. To address this limitation, a microphysiological system (MPS) is established to examine how varying levels of shear stress impact cartilage homeostasis. The system allows for the cultivation of 3D chondrogenic microconstructs (CMCs) derived from human mesenchymal stromal cells, simulating both physiological and pathophysiological shear stress. Inflammation is induced via TNF-α or activated peripheral blood mononuclear cells to model cartilage damage, enabling the evaluation of therapeutic interventions. The study demonstrates the development of an arthritis-like phenotype and successful restoration of cartilage conditions through a JAK inhibitor under physiological shear stress. Physiological shear stress is identified as a critical factor in maintaining cartilage integrity. This MPS offers a standardized method to study shear stress, replicate cytokine-induced cartilage damage, and simulate key features of arthritis, providing a valuable alternative to animal models.
Keywords: TNF‐α; articular cartilage breakdown; chondrogenic 3D model; immune cells; matrix degeneration; mesenchymal stromal cell; microfluidic system; pro‐inflammatory cytokine.
© 2024 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
M. Herrmann and T. Leeuw are employees of Sanofi‐Aventis Deutschland GmbH, which partly supported the study. The other authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- a) Burmester G. R., Feist E., Dorner T., Nat. Rev. Rheumatol. 2014, 10, 77; - PubMed
- b) Smolen J. S., Aletaha D., Barton A., Burmester G. R., Emery P., Firestein G. S., Kavanaugh A., McInnes I. B., Solomon D. H., Strand V., Yamamoto K., Nat. Rev. Dis. Primers 2018, 4, 18001; - PubMed
- c) Iannone F., Lapadula G., Aging Clin. Exp. Res. 2003, 15, 364. - PubMed
-
- a) Firestein G. S., McInnes I. B., Immunity 2017, 46, 183; - PMC - PubMed
- b) Sanchez‐Lopez E., Coras R., Torres A., Lane N. E., Guma M., Nat. Rev. Rheumatol. 2022, 18, 258; - PMC - PubMed
- c) Glyn‐Jones S., Palmer A. J., Agricola R., Price A. J., Vincent T. L., Weinans H., Carr A. J., Lancet 2015, 386, 376. - PubMed
-
- Maglaviceanu A., Wu B., Kapoor M., Wound Repair Regen. 2021, 29, 642. - PubMed
-
- Vincent T. L., Curr. Opin. Pharmacol. 2013, 13, 449. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
