Harnessing the TAF1 Acetyltransferase for Targeted Acetylation of the Tumor Suppressor p53
- PMID: 39716936
- PMCID: PMC11831463
- DOI: 10.1002/advs.202413377
Harnessing the TAF1 Acetyltransferase for Targeted Acetylation of the Tumor Suppressor p53
Abstract
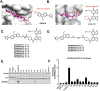
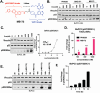
Pharmacological reactivation of the tumor suppressor p53 remains a key challenge for the treatment of cancer. Acetylation Targeting Chimera (AceTAC), a novel technology is previously reported that hijacks lysine acetyltransferases p300/CBP to acetylate the p53Y220C mutant. However, p300/CBP are the only acetyltransferases harnessed for AceTAC development to date. In this study, it is demonstrated for the first time that the TAF1 acetyltransferase can be recruited to acetylate p53Y220C. A novel TAF1-recruiting AceTAC, MS172 is discovered, which effectively acetylates p53Y220C lysine 382 in a concentration-, time- and TAF1-dependent manner via inducing the ternary complex formation between p53Y220C and TAF1. Notably, MS172 suppresses the proliferation in multiple p53Y220C-harboring cancer cell lines more potently than the previously reported p300/CBP-recruiting p53Y220C AceTAC MS78 with little toxicity in p53 WT and normal cells. Additionally, MS172 is bioavailable in mice and suitable for in vivo efficacy studies. Lastly, novel upregulation of metallothionine proteins by MS172-induced p53Y220C acetylation is discovered using RNA-seq and RT-qPCR studies. This work demonstrates that TAF1 can be harnessed for AceTAC development and expands the very limited repertoire of the acetyltransferases that can be leveraged for developing AceTACs, thus advancing the targeted protein acetylation field.
Keywords: TAF1; acetac; p53y220c; targeted protein acetylation.
© 2024 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare the following competing financial interest(s): J.J. is a cofounder and equity shareholder in Cullgen, Inc., a scientific cofounder and scientific advisory board member of Onsero Therapeutics, Inc., and a consultant for Cullgen, Inc., EpiCypher, Inc., Accent Therapeutics, Inc, and Tavotek Biotherapeutics, Inc. The Jin laboratory received research funds from Celgene Corporation, Levo Therapeutics, Inc., Cullgen, Inc. and Cullinan Oncology, Inc.
Figures



References
MeSH terms
Substances
Grants and funding
- T32CA078207/Post-doctoral Training Grant in Cancer Biology
- T32 CA078207/CA/NCI NIH HHS/United States
- UL1 TR004419/TR/NCATS NIH HHS/United States
- T32CA078207/NCI-funded pre-doctoral training grant in Cancer Biology
- Icahn School of Medicine at Mount Sinai
- Scientific Computing and Data
- S10 OD028504/OD/NIH HHS/United States
- R01 CA267696/CA/NCI NIH HHS/United States
- 1S10OD025132/Foundation for the National Institutes of Health
- R35 CA220491/CA/NCI NIH HHS/United States
- 1S10OD028504/Foundation for the National Institutes of Health
- R01 CA271346/CA/NCI NIH HHS/United States
- S10 OD025132/OD/NIH HHS/United States
- R01 CA237264/CA/NCI NIH HHS/United States
- UL1TR004419/Clinical and Translational Science Awards
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
