Light-Activatable Ubiquitin for Studying Linkage-Specific Ubiquitin Chain Formation Kinetics
- PMID: 39716962
- PMCID: PMC11809417
- DOI: 10.1002/advs.202406570
Light-Activatable Ubiquitin for Studying Linkage-Specific Ubiquitin Chain Formation Kinetics
Abstract
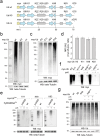
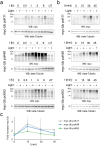
Ubiquitination is a dynamic post-translational modification governing protein abundance, function, and localization in eukaryotes. The Ubiquitin protein is conjugated to lysine residues of target proteins, but can also repeatedly be ubiquitinated itself, giving rise to a complex code of ubiquitin chains with different linkage types. To enable studying the cellular dynamics of linkage-specific ubiquitination, light-activatable polyubiquitin chain formation is reported here. By incorporating a photocaged lysine at specific sites within ubiquitin through amber codon suppression, light-dependent activation of ubiquitin chain extension is enabled for the monitoring of linkage-specific polyubiquitination. The studies reveal rapid, minute-scale ubiquitination kinetics for K11, K48, and K63 linkages. The role of individual components of the ubiquitin-proteasome system in K48-initiated chain synthesis is further studied by small molecule inhibition. The approach expands current perturbation strategies with the ability to control linkage-specific ubiquitination with high temporal resolution and should find broad application for studying ubiquitinome dynamics.
Keywords: genetic code expansion; optochemical biology; small molecule inhibitors; ubiquitin.
© 2024 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
