Exploring the potential role of ENPP2 in polycystic ovary syndrome and endometrial cancer through bioinformatic analysis
- PMID: 39717045
- PMCID: PMC11665432
- DOI: 10.7717/peerj.18666
Exploring the potential role of ENPP2 in polycystic ovary syndrome and endometrial cancer through bioinformatic analysis
Abstract
Background: Growing evidence indicates a significant correlation between polycystic ovary syndrome (PCOS) and endometrial carcinoma (EC); nevertheless, the fundamental molecular mechanisms involved continue to be unclear.
Methods: Initially, differential analysis, the least absolute shrinkage and selection operator (LASSO) regression, and support vector machine-recursive feature elimination (SVM-RFE) algorithms were employed to identify candidate genes associated with ferroptosis in PCOS. Subsequently, the TCGA-UCEC data were utilized to pinpoint the core gene. Then, the expression of ENPP2 in granulosa cells and endometrium of PCOS was validated using real-time PCR (RT-qPCR). Additionally, we investigated the role of ENPP2 in the progression from PCOS to EC through western blotting (WB), colony formation assay, cell scratch assay, transwell assay, and immunofluorescence (IF). Subsequently, ENPP2 gene set enrichment analysis (GSEA) analyses were conducted to identify common pathways involved in PCOS and EC, which were then verified by RT-qPCR. Finally, immune infiltration and the tumor microenvironment (TME) were explored to examine the involvement of ENPP2 in EC progression.
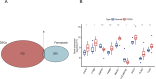
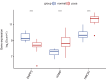
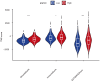
Results: The datasets TCGA-UCEC (pertaining to EC), GSE34526, GSE137684, and GSE6798 (related to PCOS) were procured and subjected to analysis. The gene ENPP2 has been recognized as the shared element connecting PCOS and EC. Next, we observed a significant downregulation of ENPP2 expression in the granulosa cells in PCOS compared to the normal patients, while an upregulation of ENPP2 expression was observed in the endometrium of hyperandrogenic PCOS patients relative to the normal. In vitro, the WB revealed that 5-dihydrotestosterone (DHT) upregulated ENPP2 expression in Ishikawa and HEC-1-A cells. Additionally, we found that ENPP2 promoted the proliferation, migration, and invasion of Ishikawa and HEC-1-A cells. Subsequently, we discovered that overexpressed ENPP2 may lead to an increase in CYP19A1 (aromatase) and AR mRNA level. IF demonstrated that ENPP2 increased the expression of AR, suggesting a regulatory role for ENPP2 in hormonal response within PCOS and EC. Our findings indicated a significant correlation between ENPP2 expression and the modulation of immune responses.
Keywords: Bioinformatic analysis; ENPP2; Endometrial cancer; Ferroptosis; Polycystic ovary syndrome.
© 2024 Zhang et al.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures








References
-
- Auciello FR, Bulusu V, Oon C, Tait-Mulder J, Berry M, Bhattacharyya S, Tumanov S, Allen-Petersen BL, Link J, Kendsersky ND, Vringer E, Schug M, Novo D, Hwang RF, Evans RM, Nixon C, Dorrell C, Morton JP, Norman JC, Sears RC, Kamphorst JJ, Sherman MH. A stromal lysolipid-autotaxin signaling axis promotes pancreatic tumor progression. Cancer Discovery. 2019;9(5):617–627. doi: 10.1158/2159-8290.CD-18-1212. - DOI - PMC - PubMed
-
- Carter CA, Pogribny M, Davidson A, Jackson CD, McGarrity LJ, Morris SM. Effects of retinoic acid on cell differentiation and reversion toward normal in human endometrial adenocarcinoma (RL95-2) cells. Anticancer Research. 1996;16:17–24. - PubMed
-
- Chen SU, Lee H, Chang DY, Chou CH, Chang CY, Chao KH, Lin CW, Yang YS. Lysophosphatidic acid mediates interleukin-8 expression in human endometrial stromal cells through its receptor and nuclear factor-kappaB-dependent pathway: a possible role in angiogenesis of endometrium and placenta. Endocrinology. 2008;149(11):5888–5896. doi: 10.1210/en.2008-0314. - DOI - PubMed
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

