Bioinformatics Analysis of Biomarkers and Therapeutic Targets Related to Necroptosis in Intervertebral Disc Degeneration
- PMID: 39717265
- PMCID: PMC11666314
- DOI: 10.1155/bmri/9922966
Bioinformatics Analysis of Biomarkers and Therapeutic Targets Related to Necroptosis in Intervertebral Disc Degeneration
Abstract
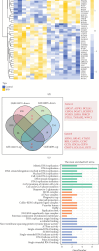
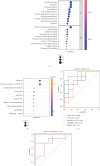
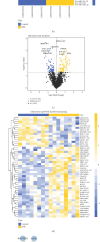
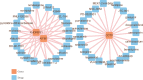
Necroptosis is a critical process in intervertebral disc degeneration (IDD). This research is aimed at identifying key genes regulating necroptosis in IDD to provide a theoretical basis for early diagnosis and treatment. Transcriptome data from patients with IDD and normal samples were obtained from the GSE34095 and GSE124272 datasets of the Gene Expression Omnibus (GEO) public database. Necroptosis-related genes (NRGs) were sourced from the GeneCards database and literature. Differentially expressed necroptosis-related genes (DE-NRGs) in IDD were identified by intersecting these sources. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) were used for gene annotation analysis. The receiver operating characteristic (ROC) curve and nomogram analyses assessed the diagnostic efficiency of DE-NRGs. The miRWalk and starBase databases helped construct the competing endogenous RNA (ceRNA) regulatory network of DE-NRGs. We identified 517 differential genes in tissue and 2974 in blood, with 62 genes in common. DE-NRGs (AIFM1, CCT8, HNRNPA1, KHDRBS1, SERBP1) were identified by intersecting NRGs with these 62 common genes. The ROC curve showed an area under the curve (AUC) > 0.70 for DE-NRGs, and the nomogram indicated that a higher DE-NRG score correlates with a higher risk of IDD. CCT8, KHDRBS1, and AIFM1 emerged as potential therapeutic targets for IDD through target drug prediction. qRT-PCR (quantitative reverse transcription polymerase chain reaction), Western blot, and immunohistochemistry confirmed the expression of AIFM1, CCT8, HNRNPA1, KHDRBS1, and SERBP1 in patients' nucleus pulposus tissue, suggesting these genes as key targets for IDD risk assessment and drug therapy.
Keywords: ceRNA network; diagnostic markers; immune infiltration; intervertebral disc degeneration; necroptosis; therapeutic target.
Copyright © 2024 Fan Zhang et al.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Similar articles
-
Integrating bioinformatics and experimental validation to Investigate IRF1 as a novel biomarker for nucleus pulposus cells necroptosis in intervertebral disc degeneration.Sci Rep. 2024 Dec 3;14(1):30138. doi: 10.1038/s41598-024-81681-8. Sci Rep. 2024. PMID: 39627301 Free PMC article.
-
Exploring the molecular mechanisms underlying intervertebral disc degeneration by analysing multiple datasets.Sci Rep. 2025 Apr 28;15(1):14748. doi: 10.1038/s41598-025-98070-4. Sci Rep. 2025. PMID: 40289127 Free PMC article.
-
Bioinformatics Research and qRT-PCR Verify Hub Genes and a Transcription Factor-MicroRNA Feedback Network in Intervertebral Disc Degeneration.Appl Biochem Biotechnol. 2024 Jun;196(6):3184-3198. doi: 10.1007/s12010-023-04699-0. Epub 2023 Aug 26. Appl Biochem Biotechnol. 2024. PMID: 37632659
-
Combined single-cell RNA sequencing and mendelian randomization to identify biomarkers associated with necrotic apoptosis in intervertebral disc degeneration.Spine J. 2025 Jan;25(1):165-183. doi: 10.1016/j.spinee.2024.09.011. Epub 2024 Sep 25. Spine J. 2025. PMID: 39332686
-
ATG9A as a potential diagnostic marker of intervertebral disc degeneration: Inferences from experiments and bioinformatics analysis incorporating sc-RNA-seq data.Gene. 2024 Mar 1;897:148084. doi: 10.1016/j.gene.2023.148084. Epub 2023 Dec 15. Gene. 2024. PMID: 38104954
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

