Elucidating metabolic pathways through genomic analysis in highly heavy metal-resistant Halobacterium salinarum strains
- PMID: 39717611
- PMCID: PMC11665356
- DOI: 10.1016/j.heliyon.2024.e40822
Elucidating metabolic pathways through genomic analysis in highly heavy metal-resistant Halobacterium salinarum strains
Abstract
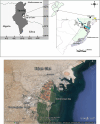
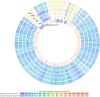
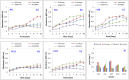
The annotated and predicted genomes of five archaeal strains (AS1, AS2, AS8, AS11 and AS19), isolated from Sfax solar saltern sediments (Tunisia) and affiliated with Halobacterium salinarum, were performed by RAST webserver (Rapid Annotation using Subsystem Technology) and NCBI prokaryotic genome annotation pipeline (PGAP). The results showed the ability of strains to use a reduced semi-phosphorylative Entner-Doudoroff pathway for glucose degradation and an Embden-Meyerhof one for gluconeogenesis. They could use glucose, fructose, glycerol, and acetate as sole source of carbon and energy. ATP synthase, various cytochromes and aerobic respiration proteins were encoded. All strains showed fermentation capability through the arginine deiminase pathway and facultative anaerobic respiration using electron acceptors (Dimethyl sulfoxide and trimethylamine N-oxide). Several biosynthesis pathways for many amino acids were identified. Comparative and pangenome analyses between the strains and the well-studied halophilic archaea Halobacterium NRC-1 highlighted a notable dissimilarity. Besides, the strains shared a core genome of 1973 genes and an accessory genome of 767 genes. 129, 94, 67, 15 and 29 unique genes were detected in the AS1, AS2, AS8, AS11 and AS19 genomes, respectively. Most of these unique genes code for hypothetical proteins. The strains displayed plant-growth promoting characteristics under heavy metal stress (Ammonium assimilation, phosphate solubilization, chemotaxis, cell motility and production of indole acetic acid, siderophore and phenazine). Therefore, they could be used as a biofertilizer to promote plant growth. The genomes encoded numerous biotechnologically relevant genes responsible for vitamin biosynthesis, including cobalamin, folate, biotin, pantothenate, riboflavin, thiamine, menaquinone, nicotinate, and nicotinamide. The carotenogenetic pathway of the studied strains was also predicted. Consequently, the findings of this study contribute to a better understanding of the halophilic archaea metabolism providing valuable insights into their ecophysiology as well as relevant biotechnological applications.
Keywords: Biotechnological applications; Comparative genomics; Halobacterium salinarum; Halophilic archaea; Metabolism.
© 2024 The Authors. Published by Elsevier Ltd.
Conflict of interest statement
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Rosa martinez reports financial support was provided by 10.13039/100009092University of Alicante. Rosa martinez reports a relationship with 10.13039/100009092University of Alicante that includes: funding grants. Rosa martinez has patent licensed to PROMETEO/2021/055/Generalirat Valenciana VIGROB-309/University of Alicante. nothing If there are other authors, they declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


References
-
- Oren A. Kluwer Academic Publishers; The Netherlands: 2002. Halophilic Microorganisms and Their Environments, Volume 5 of Cellular Origin and Life in Extreme Habitats. Dordrecht.
LinkOut - more resources
Full Text Sources

