Rationalizing the effects of RNA modifications on protein interactions
- PMID: 39717617
- PMCID: PMC11664407
- DOI: 10.1016/j.omtn.2024.102391
Rationalizing the effects of RNA modifications on protein interactions
Abstract
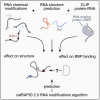
RNA modifications play a crucial role in regulating gene expression by altering RNA structure and modulating interactions with RNA-binding proteins (RBPs). In this study, we explore the impact of specific RNA chemical modifications-N6-methyladenosine (m⁶A), A-to-I editing, and pseudouridine (Ψ)-on RNA secondary structure and protein-RNA interactions. Utilizing genome-wide data, including RNA secondary structure predictions and protein-RNA interaction datasets, we classify proteins into distinct categories based on their binding behaviors: modification specific and structure independent, or modification unspecific and structure dependent. For instance, m⁶A readers such as YTHDF2 exhibit modification-specific and structure-independent binding, consistently recognizing m⁶A regardless of structural changes. Conversely, proteins such as U2AF2 display modification-unspecific and structure-dependent behavior, altering their binding preferences in response to structural changes induced by different modifications. A-to-I editing, which causes significant structural changes, typically reduces protein interactions, while Ψ enhances RNA structural stability, albeit with variable effects on protein binding. To predict these interactions, we developed the catRAPID 2.2 RNA modifications algorithm, which computes the effects of RNA modifications on protein-RNA binding propensities. This algorithm enables the prediction and analysis of RNA modifications' impact on protein interactions, offering new insights into RNA biology and engineering.
Keywords: A-to-I; MT: RNA and epigenetic editing Special Issue; RNA modifications; RNA secondary structure; RNA-binding proteins; interactions predictions; m6A; machine-learning; protein-RNA interactions; pseudouridine.
© 2024 Published by Elsevier Inc. on behalf of The American Society of Gene and Cell Therapy.
Conflict of interest statement
The authors declare no competing interests.
Figures






References
-
- Fagre C., Gilbert W. Beyond reader proteins: RNA binding proteins and RNA modifications in conversation to regulate gene expression. Wiley Interdiscip. Rev. RNA. 2024;15 - PubMed
-
- Zhou H., Kimsey I.J., Nikolova E.N., Sathyamoorthy B., Grazioli G., McSally J., Bai T., Wunderlich C.H., Kreutz C., Andricioaei I., Al-Hashimi H.M. m(1)A and m(1)G disrupt A-RNA structure through the intrinsic instability of Hoogsteen base pairs. Nat. Struct. Mol. Biol. 2016;23:803–810. - PMC - PubMed
LinkOut - more resources
Full Text Sources

