Machine Learning and Mendelian Randomization Reveal Molecular Mechanisms and Causal Relationships of Immune-Related Biomarkers in Periodontitis
- PMID: 39717623
- PMCID: PMC11666315
- DOI: 10.1155/mi/9983323
Machine Learning and Mendelian Randomization Reveal Molecular Mechanisms and Causal Relationships of Immune-Related Biomarkers in Periodontitis
Abstract
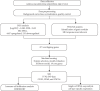
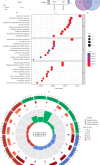
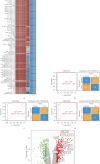
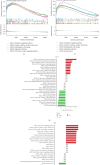
This study aimed to investigate the molecular mechanisms of periodontitis and identify key immune-related biomarkers using machine learning and Mendelian randomization (MR). Differentially expressed gene (DEG) analysis was performed on periodontitis datasets GSE16134 and GSE10334 from the Gene Expression Omnibus (GEO) database, followed by weighted gene co-expression network analysis (WGCNA) to identify relevant gene modules. Various machine learning algorithms were utilized to construct predictive models, highlighting core genes, while MR assessed the causal relationships between these genes and periodontitis. Additionally, immune infiltration analysis and single-cell sequencing were employed to explore the roles of key genes in immunity and their expression across different cell types. The integration of machine learning, MR, and single-cell sequencing represents a novel approach that significantly enhances our understanding of the immune dynamics and gene interactions in periodontitis. The study identified 682 significant DEGs, with WGCNA revealing seven gene modules associated with periodontitis and 471 core candidate genes. Among the 113 machine learning algorithms tested, XGBoost was the most effective in identifying periodontitis samples, leading to the selection of 19 core genes. MR confirmed significant causal relationships between CD93, CD69, and CXCL6 and periodontitis. Further analysis showed that these genes were correlated with various immune cells and exhibited specific expression patterns in periodontitis tissues. The findings suggest that CD93, CD69, and CXCL6 are closely related to the progression of periodontitis, with MR confirming their causal links to the disease. These genes have potential applications in the diagnosis and treatment of periodontitis, offering new insights into the disease's molecular mechanisms and providing valuable resources for precision medicine approaches in periodontitis management. Limitations of this study include the demographic and sample size constraints of the datasets, which may impact the generalizability of the findings. Future research is needed to validate these biomarkers in larger, diverse cohorts and to investigate their functional roles in the pathogenesis of periodontitis.
Keywords: Mendelian randomization; biomarkers; machine learning; periodontitis; single-cell sequencing.
Copyright © 2024 Yuan Li et al.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


Similar articles
-
Screening COPD-Related Biomarkers and Traditional Chinese Medicine Prediction Based on Bioinformatics and Machine Learning.Int J Chron Obstruct Pulmon Dis. 2024 Sep 24;19:2073-2095. doi: 10.2147/COPD.S476808. eCollection 2024. Int J Chron Obstruct Pulmon Dis. 2024. PMID: 39346628 Free PMC article.
-
Exploration of the shared diagnostic genes and mechanisms between periodontitis and primary Sjögren's syndrome by integrated comprehensive bioinformatics analysis and machine learning.Int Immunopharmacol. 2024 Nov 15;141:112899. doi: 10.1016/j.intimp.2024.112899. Epub 2024 Aug 13. Int Immunopharmacol. 2024. PMID: 39142001
-
Identification and Validation of Ferritinophagy-Related Biomarkers in Periodontitis.Int Dent J. 2025 Jun;75(3):1781-1797. doi: 10.1016/j.identj.2025.03.011. Epub 2025 Apr 15. Int Dent J. 2025. PMID: 40233623 Free PMC article.
-
Deciphering the role of pyroptosis-related genes and natural killer T cells in sepsis pathogenesis: a comprehensive bioinformatics and Mendelian randomization analysis.J Physiol Pharmacol. 2025 Apr;76(2). doi: 10.26402/jpp.2025.2.10. Epub 2025 May 5. J Physiol Pharmacol. 2025. PMID: 40350654
-
Exploring the potential causal relationship between gut microbiota and heart failure: A two-sample mendelian randomization study combined with the geo database.Curr Probl Cardiol. 2024 Feb;49(2):102235. doi: 10.1016/j.cpcardiol.2023.102235. Epub 2023 Nov 30. Curr Probl Cardiol. 2024. PMID: 38040216 Review.
Cited by
-
The Transformative Role of Artificial Intelligence in Dentistry: A Comprehensive Overview. Part 1: Fundamentals of AI, and its Contemporary Applications in Dentistry.Int Dent J. 2025 Apr;75(2):383-396. doi: 10.1016/j.identj.2025.02.005. Epub 2025 Mar 11. Int Dent J. 2025. PMID: 40074616 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

