Diaporthe species on palms - integrative taxonomic approach for species boundaries delimitation in the genus Diaporthe, with the description of D. pygmaeae sp. nov
- PMID: 39717652
- PMCID: PMC11663421
- DOI: 10.3114/sim.2024.109.08
Diaporthe species on palms - integrative taxonomic approach for species boundaries delimitation in the genus Diaporthe, with the description of D. pygmaeae sp. nov
Abstract
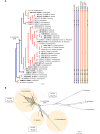
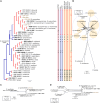
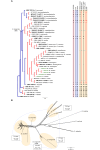
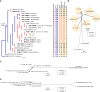
The application of traditional morphological and ecological species concepts to closely related, asexual fungal taxa is challenging due to the lack of distinctive morphological characters and frequent cosmopolitan and plurivorous behaviour. As a result, multilocus sequence analysis (MLSA) has become a powerful and widely used tool to recognise and delimit independent evolutionary lineages (IEL) in fungi. However, MLSA can mask discordances in individual gene trees and lead to misinterpretation of speciation events. This phenomenon has been extensively documented in Diaporthe, and species identifications in this genus remains an ongoing challenge. However, the accurate delimitation of Diaporthe species is critical as the genus encompasses several cosmopolitan pathogens that cause serious diseases on many economically important plant hosts. In this regard, following a survey of palm leaf spotting fungi in Lisbon, Portugal, Diaporthe species occurring on Arecaceae hosts were used as a case study to implement an integrative taxonomic approach for a reliable species identification in the genus. Molecular analyses based on the genealogical concordance phylogenetic species recognition (GCPSR) and DNA-based species delimitation methods revealed that speciation events in the genus have been highly overestimated. Most IEL identified by the GCPSR were also recognised by Poisson tree processes (PTP) coalescent-based methods, which indicated that phylogenetic lineages in Diaporthe are likely influenced by incomplete lineage sorting (ILS) and reticulation events. Furthermore, the recognition of genetic recombination signals and the evaluation of genetic variability based on sequence polymorphisms reinforced these hypotheses. New clues towards the intraspecific variation in the common loci used for phylogenetic inference of Diaporthe species are discussed. These results demonstrate that intraspecific variability has often been used as an indicator to introduce new species in Diaporthe, which has led to a proliferation of species names in the genus. Based on these data, 53 species are reduced to synonymy with 18 existing Diaporthe species, and a new species, D. pygmaeae, is introduced. Thirteen new plant host-fungus associations are reported, all of which represent new host family records for Arecaceae. This study has recognised and resolved a total of 14 valid Diaporthe species associated with Arecaceae hosts worldwide, some of which are associated with disease symptoms. This illustrates the need for more systematic research to examine the complex of Diaporthe taxa associated with palms and determine their potential pathogenicity. By implementing a more rational framework for future studies on species delimitation in Diaporthe, this study provides a solid foundation to stabilise the taxonomy of species in the genus. Guidelines for species recognition, definition and identification in Diaporthe are included. Taxonomic novelties: New species: Diaporthe pygmaeae D.S. Pereira & A.J.L. Phillips. New synonyms: Diaporthe afzeliae Monkai & Lumyong, Diaporthe alangii C.M. Tian & Q. Yang, Diaporthe araliae-chinensis S.Y. Wang et al., Diaporthe australiana R.G. Shivas et al., Diaporthe australpacifica Y.P. Tan & R.G. Shivas, Diaporthe bombacis Monkai & Lumyong, Diaporthe caryae C.M. Tian & Q. Yang, Diaporthe chimonanthi (C.Q. Chang et al.) Y.H. Gao & L. Cai, Diaporthe conferta H. Dong et al., Diaporthe diospyrina Y.K. Bai & X.L. Fan, Diaporthe durionigena L.D. Thao et al., Diaporthe etinsideae Y.P. Tan & R.G. Shivas, Diaporthe eucalyptorum Crous & R.G. Shivas, Diaporthe fujianensis Jayaward. et al., Diaporthe fusiformis Jayaward. et al., Diaporthe globoostiolata Monkai & Lumyong, Diaporthe hainanensis Qin Yang, Diaporthe hongkongensis R.R. Gomes et al., Diaporthe hubeiensis Dissan. et al., Diaporthe infecunda R.R. Gomes et al., Diaporthe italiana Chethana et al., Diaporthe juglandigena S.Y. Wang et al., Diaporthe lagerstroemiae (C.Q. Chang et al.) Y.H. Gao & L. Cai, Diaporthe lithocarpi (Y.H. Gao et al.) Y.H. Gao & L. Cai, Diaporthe lutescens S.T. Huang et al., Diaporthe machili S.T. Huang et al., Diaporthe megabiguttulata M. Luo et al., Diaporthe middletonii R.G. Shivas et al., Diaporthe morindae M. Luo et al., Diaporthe nannuoshanensis S.T. Huang et al., Diaporthe nigra Brahman. & K.D. Hyde, Diaporthe orixae Q.T. Lu & Zhen Zhang, Diaporthe passifloricola Crous & M.J. Wingf., Diaporthe pimpinellae Abeywickrama et al., Diaporthe pseudoinconspicua T.G.L Oliveira et al., Diaporthe pungensis S.T. Huang et al., Diaporthe rhodomyrti C.M. Tian & Qin Yang, Diaporthe rosae M.C. Samar. & K.D. Hyde, Diaporthe rumicicola Manawas et al., Diaporthe salicicola R.G. Shivas et al., Diaporthe samaneae Monkai & Lumyong, Diaporthe subcylindrospora S.K. Huang et al., Diaporthe tectonae Doilom et al., Diaporthe tectonigena Doilom et al., Diaporthe theobromatis H. Dong et al., Diaporthe thunbergiicola Udayanga & K.D. Hyde, Diaporthe tuyouyouiae Y.P. Tan et al., Diaporthe unshiuensis F. Huang et al., Diaporthe vochysiae S.A. Noriler et al., Diaporthe xishuangbannaensis Hongsanan & K.D. Hyde, Diaporthe xylocarpi M.S. Calabon & E.B.G. Jones, Diaporthe zaobaisu Y.S. Guo & G.P. Wang, Diaporthe zhaoqingensis M. Luo et al. Citation: Pereira DS, Phillips AJL (2024). Diaporthe species on palms - integrative taxonomic approach for species boundaries delimitation in the genus Diaporthe, with the description of D. pygmaeae sp. nov. Studies in Mycology 109: 487-594. doi: 10.3114/sim.2024.109.08.
Keywords: coalescent-based methods; genetic distance-based methods; integrative taxonomy; new taxa; species limits.
© 2024 Westerdijk Fungal Biodiversity Institute.
Conflict of interest statement
The authors declare that there is no conflict of interest.
Figures
















References
-
- Adaskaveg JE, Förster H, Connell JH. (1999). First report of fruit rot and associated branch dieback of almond in California caused by a Phomopsis species tentatively identified as P. amygdali. Plant Disease 83: 1073. - PubMed
-
- Ahrens D, Fujisawa T, Krammer H-J, et al. (2016). Rarity and incomplete sampling in DNA-based species delimitation. Systematic Biology 65: 478–494. - PubMed
-
- Aiello D, Guarnaccia V, Costanzo MB. et al. (2022). Woody canker and shoot blight caused by Botryosphaeriaceae and Diaporthaceae on mango and litchi in Italy. Horticulturae 8: 330.
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
