Proposal of one new family, seven new genera and seventy new basidiomycetous yeast species mostly isolated from Tibet and Yunnan provinces, China
- PMID: 39717653
- PMCID: PMC11663428
- DOI: 10.3114/sim.2024.109.02
Proposal of one new family, seven new genera and seventy new basidiomycetous yeast species mostly isolated from Tibet and Yunnan provinces, China
Abstract
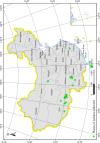
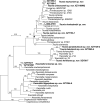
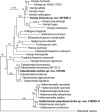
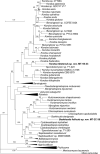
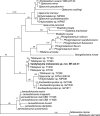
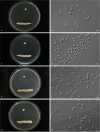
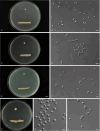
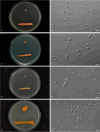
More than 2 000 yeast strains isolated from 1 200 samples mostly collected from Tibet and Yunnan provinces in China were identified as 462 species according to the internal transcribed spacer including the 5.8S rDNA (ITS) and the D1/D2 domains of the large subunit rDNA (LSU) sequence analyses. Among them, 70 new basidiomycetous yeast species were proposed based on the multi-locus phylogenetic analyses including the D1/D2 domains, the ITS, the small subunit rDNA (SSU), the largest subunit of RNA polymerase II (RPB1), the second largest subunit of RNA polymerase II (RPB2) and translation elongation factor 1-α (TEF1), as well as the phenotypic comparisons. The average nucleotide identity (ANI) analysis with the genomic metric was also used in the evaluation of the species delimitation for the genera Holtermannia, Mrakia and Takashimella that contain closely related species with low sequence heterogeneity in both ITS and D1/D2 regions. Forty-six new species belonged to 16 genera in the Agaricomycotina, 13 new species occurred in 12 genera in the Pucciniomycotina, three new species were distributed in three genera in the Ustilaginomycotina, and eight new species were classified in seven newly established genera. One new family was also proposed based on one of these new genera. The analyses revealed several inaccurate species names attributed to genomes deposited in GenBank, indicating the necessity of a more rigorous quality checks of the genomes deposited in the public databases. Taxonomic novelties: New family: Vankyiozymaceae Q.-M. Wang; New genera: Baiomyces Q.-M. Wang, Bauerozyma Q.-M. Wang, Fengyania Q.-M. Wang, Foliozyma Q.-M. Wang, Litoriozyma Q.-M. Wang, Nakaseozyma Q.-M. Wang, Vankyiozyma Q.-M. Wang; New species: Baiomyces sejilaensis Q.-M. Wang, Bauerozyma artemisiae Q.-M. Wang, Boekhoutia foliicola Q.-M. Wang, Buckleyzyma pseudoaurantiaca Q.-M. Wang, Carlosrosaea betulae Q.-M. Wang, Carlosrosaea rhododendri Q.-M. Wang, Carlosrosaea yunnanensis Q.-M. Wang, Chrysozyma quercicola Q.-M. Wang, Curvibasidium pini Q.-M. Wang, Cystobasidium cunninghamiae Q.-M. Wang, Derxomyces foliicola Q.-M. Wang, Derxomyces motuoensis Q.-M. Wang, Derxomyces orientalis Q.-M. Wang, Derxomyces paracylindricus Q.-M. Wang, Exobasidium lijiangense Q.-M. Wang, Fengyania pteridophytorum Q.-M. Wang, Foliozyma liliicola Q.-M. Wang, Halobasidium phyllophilum Q.-M. Wang, Hannaella artemisiae Q.-M. Wang, Hannaella pteridophytorum Q.-M. Wang, Hannaella urticae Q.-M. Wang, Holtermannia pseudosaccardoi Q.-M. Wang, Kockovaella cariosiligni Q.-M. Wang, Kockovaella foliicola Q.-M. Wang, Kondoa tibetensis Q.-M. Wang, Kwoniella hippophaes Q.-M. Wang, Kwoniella lonicerae Q.-M. Wang, Litoriozyma hainanensis Q.-M. Wang, Meira marina Q.-M. Wang, Microsporomyces betulae Q.-M. Wang, Microsporomyces foliicola Q.-M. Wang, Mrakia pini Q.-M. Wang, Mrakia rhododendri Q.-M. Wang, Nakaseozyma junci Q.-M.Wang, Nakaseozyma lonicerae Q.-M. Wang, Papiliotrema castaneae Q.-M. Wang, Papiliotrema catalpae Q.-M. Wang, Phaeotremella pini Q.-M. Wang, Phaffia paratasmanica Q.-M. Wang, Phaffia rhododendri Q.-M. Wang, Piskurozyma cuscutae Q.-M. Wang, Piskurozyma humicola Q.-M. Wang, Piskurozyma liliaceifoliae Q.-M. Wang, Piskurozyma linzhiensis Q.-M. Wang, Piskurozyma nanyiensis Q.-M. Wang, Piskurozyma terricola Q.-M. Wang, Pseudohyphozyma sanghuangpori Q.-M. Wang, Pseudotremella hippophaes Q.-M. Wang, Pseudotremella rhododendri Q.-M. Wang, Rhodotorula linzhiensis Q.-M. Wang, Slooffia terricola Q.-M. Wang, Takashimella corticis Q.-M. Wang, Teunia betulicola Q.-M. Wang, Teunia chimonanthi Q.-M. Wang, Teunia heritierae Q.-M. Wang, Teunia myricariae Q.-M. Wang, Teunia parabetulicola Q.-M. Wang, Teunia quercus Q.-M. Wang, Teunia rhododendri Q.-M. Wang, Ustilago foliicola Q.-M. Wang, Vankyiozyma motuoensis Q.-M. Wang, Vanrija silvicola Q.-M. Wang, Vishniacozyma catalpae Q.-M. Wang, Vishniacozyma marinae Q.-M. Wang, Vishniacozyma paravictoriae Q.-M. Wang, Vishniacozyma pini Q.-M. Wang, Vishniacozyma pyri Q.-M. Wang, Vishniacozyma sinopodophylli Q.-M. Wang, Vishniacozyma zhenxiongensis Q.-M. Wang, Yurkovia castaneae Q.-M. Wang. Citation: Jiang Y-L, Bao W-J, Liu F, Wang G-S, Yurkov AM, Ma Q, Hu Z-D, Chen X-H, Zhao W-N, Li A-H, Wang Q-M (2024). Proposal of one new family, seven new genera and seventy new basidiomycetous yeast species mostly isolated from Tibet and Yunnan provinces, China. Studies in Mycology 109: 57-153. doi: 10.3114/sim.2024.109.02.
Keywords: Average nucleotide identity (ANI); basidiomycetous yeasts; genomic metrics; molecular phylogeny; seventy-eight new taxa.
© 2024 Westerdijk Fungal Biodiversity Institute.
Conflict of interest statement
The authors declare that there is no conflict of interest.
Figures



































Similar articles
-
Diversity and phylogeny of basidiomycetous yeasts from plant leaves and soil: Proposal of two new orders, three new families, eight new genera and one hundred and seven new species.Stud Mycol. 2020 Jan 28;96:17-140. doi: 10.1016/j.simyco.2020.01.002. eCollection 2020 Jun. Stud Mycol. 2020. PMID: 32206137 Free PMC article.
-
Towards an integrated phylogenetic classification of the Tremellomycetes.Stud Mycol. 2015 Jun;81:85-147. doi: 10.1016/j.simyco.2015.12.001. Epub 2016 Jan 8. Stud Mycol. 2015. PMID: 26955199 Free PMC article.
-
Phylogenetic classification of yeasts and related taxa within Pucciniomycotina.Stud Mycol. 2015 Jun;81:149-89. doi: 10.1016/j.simyco.2015.12.002. Epub 2016 Jan 11. Stud Mycol. 2015. PMID: 26951631 Free PMC article.
-
Taxonomy, phylogeny and identification of Chaetomiaceae with emphasis on thermophilic species.Stud Mycol. 2022 Jul;101:121-243. doi: 10.3114/sim.2022.101.03. Epub 2022 Apr 1. Stud Mycol. 2022. PMID: 36059895 Free PMC article.
-
Revising Clonostachys and allied genera in Bionectriaceae.Stud Mycol. 2023 Jun;105:205-266. doi: 10.3114/sim.2023.105.03. Epub 2023 Jun 12. Stud Mycol. 2023. PMID: 38895704 Free PMC article.
Cited by
-
Three new species of Carlosrosaea (Trimorphomycetaceae, Tremellales) from China.MycoKeys. 2025 Jul 1;119:123-135. doi: 10.3897/mycokeys.119.151751. eCollection 2025. MycoKeys. 2025. PMID: 40641588 Free PMC article.
-
A new psychrophilic yeast of Kriegeriaceae (Kriegeriales) isolated from lichen in the Arctic, with the description of Licheniasvalbardensis gen. et sp. nov.MycoKeys. 2025 Feb 24;114:95-113. doi: 10.3897/mycokeys.114.135299. eCollection 2025. MycoKeys. 2025. PMID: 40041796 Free PMC article.
-
Rhodotorula silvicola sp. nov., a new yeast species from plant-associated substrates and mushroom.Int J Syst Evol Microbiol. 2025 Jul;75(7):006836. doi: 10.1099/ijsem.0.006836. Int J Syst Evol Microbiol. 2025. PMID: 40643100 Free PMC article.
-
Five new epiphytic species of Vishniacozyma (Bulleribasidiaceae, Tremellales) from China.MycoKeys. 2025 Feb 12;113:321-336. doi: 10.3897/mycokeys.113.140598. eCollection 2025. MycoKeys. 2025. PMID: 39980722 Free PMC article.
-
Two new arthroconidial yeast species from bark and pit mud in China.MycoKeys. 2025 Jan 28;113:57-72. doi: 10.3897/mycokeys.113.141799. eCollection 2025. MycoKeys. 2025. PMID: 39911477 Free PMC article.
References
-
- Begerow D, Kemler M, Feige A. et al. (2017). Parasitism in yeasts. In: Yeasts in natural ecosystems: Ecology. (Buzzini P, Lachance MA, Yurkov A, eds). Springer Cham; Switserland: 179–210.
-
- Belisle M, Mendenhall CD, Brenes FO. et al. (2014). Temporal variation in fungal communities associated with tropical hummingbirds and nectarivorous bats. Fungal Ecology 12: 44–51.
-
- Bing J, Han PJ, Liu WQ. et al. (2014). Evidence for a Far East Asian origin of lager beer yeast. Current Biology 24: R380–R381. - PubMed
Associated data
LinkOut - more resources
Full Text Sources
