Sporocadaceae revisited
- PMID: 39717655
- PMCID: PMC11663424
- DOI: 10.3114/sim.2024.109.03
Sporocadaceae revisited
Abstract
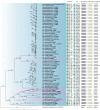
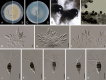
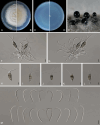
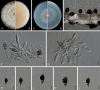
Sporocadaceae is a species-rich and cosmopolitan fungal family including species of plant pathogens, endophytes or saprobes, and parasites of humans and animals. The taxonomy of Sporocadaceae has recently been revised using a polyphasic approach. However, much remains unknown about the diversity of species and their host associations. A collection of 488 strains, mostly from China and associated with 129 host plant species, was studied based on morphological comparisons and multi-locus (LSU, ITS, tef-1α, tub2, and rpb2) phylogenies. Our results revealed that they belonged to 86 species, one new genus (Cavernicola gen. nov.) and seven known genera, including Discosia, Monochaetia, Neopestalotiopsis, Pestalotiopsis, Seimatosporium, Seiridium and Sporocadus. Of these, 43 new species and three new combinations (Dis. kaki, Mon. bulbophylli, and Neo. keteleeriae) are proposed in this paper. In addition, Neo. vaccinii, Pes. kaki and Pes. nanjingensis are synonymised under Neo. hispanica, Pes. menhaiensis and Pes. sichuanensis, respectively. We also corrected seven problematic sequences of type materials of previously published species, namely Neo. iranensis (tef-1α, ITS, tub2), Pes. jesteri (tef-1α), Pes. photinicola (ITS, tub2) and Pes. yunnanensis (ITS). Based on this study, Pestalotiopsis and Neopestalotiopsis are the most commonly detected genera within the Sporocadaceae family, associated with 84 and 70 plant species, respectively. Furthermore, considering the importance of Sporocadaceae and the fact that commonly used loci provide little valid information for species delimitation in this family, especially for Neopestalotiopsis and Pestalotiopsis, we initiated a phylogenomic project in this study. It will not only contribute to the knowledge of species boundaries but will also provide an important basis for evolutionary studies and research on secondary metabolites in Sporocadaceae. Taxonomic novelties: New genus: Cavernicola P. Razaghi, F. Liu & L. Cai. New species: Cavernicola guangxiensis P. Razaghi, F. Liu & L. Cai, Discosia ascidiata P. Razaghi, F. Liu & L. Cai, Discosia jiangxiensis P. Razaghi, F. Liu & L. Cai, Discosia navicularis P. Razaghi, F. Liu & L. Cai, Neopestalotiopsis ageratinae P. Razaghi, F. Liu & L. Cai, Neopestalotiopsis castanopsidis P. Razaghi, F. Liu & L. Cai, Neopestalotiopsis celtidis P. Razaghi, F. Liu, M. Raza & L. Cai, Neopestalotiopsis collariata P. Razaghi, F. Liu & L. Cai, Neopestalotiopsis dimorphospora P. Razaghi, F. Liu & L. Cai, Neopestalotiopsis dolichoconidiophora P. Razaghi, F. Liu & L. Cai, Neopestalotiopsis fijiensis P. Razaghi, F. Liu & L. Cai, Neopestalotiopsis fimbriata P. Razaghi, F. Liu & L. Cai, Neopestalotiopsis fuzhouensis P. Razaghi, F. Liu & L. Cai, Neopestalotiopsis guangxiensis P. Razaghi, F. Liu, M. Raza & L. Cai, Neopestalotiopsis guizhouensis P. Razaghi, F. Liu, M. Raza & L. Cai, Neopestalotiopsis jiangxiensis P. Razaghi, F. Liu & L. Cai, Neopestalotiopsis liquidambaris P. Razaghi, F. Liu & L. Cai, Neopestalotiopsis machili P. Razaghi, F. Liu & L. Cai, Neopestalotiopsis megabetaspora P. Razaghi, F. Liu, M. Raza & L. Cai, Neopestalotiopsis moniliformis P. Razaghi, F. Liu & L. Cai, Neopestalotiopsis nanningensis P. Razaghi, F. Liu, M. Raza & L. Cai, Neopestalotiopsis phyllostachydis P. Razaghi, F. Liu, M. Raza & L. Cai, Neopestalotiopsis poae P. Razaghi, F. Liu & L. Cai, Neopestalotiopsis smilacis P. Razaghi, F. Liu, M. Raza & L. Cai, Pestalotiopsis alloschemones P. Razaghi, F. Liu & L. Cai, Pestalotiopsis americana P. Razaghi, F. Liu & L. Cai, Pestalotiopsis biappendiculata P. Razaghi, F. Liu & L. Cai, Pestalotiopsis cratoxyli P. Razaghi, F. Liu, M. Raza & L. Cai, Pestalotiopsis exudata P. Razaghi, F. Liu & L. Cai, Pestalotiopsis fusiformis P. Razaghi, F. Liu & L. Cai, Pestalotiopsis ganzhouensis P. Razaghi, F. Liu & L. Cai, Pestalotiopsis leucospermi P. Razaghi, F. Liu & L. Cai, Pestalotiopsis lobata P. Razaghi, F. Liu & L. Cai, Pestalotiopsis machili P. Razaghi, F. Liu & L. Cai, Pestalotiopsis multiappendiculata P. Razaghi, F. Liu & L. Cai, Pestalotiopsis pruni P. Razaghi, F. Liu & L. Cai, Pestalotiopsis rubrae P. Razaghi, F. Liu, M. Raza & L. Cai, Pestalotiopsis wulichongensis P. Razaghi, F. Liu, M. Raza & L. Cai, Seimatosporium tibetense P. Razaghi, F. Liu & L. Cai, Seiridium rhododendri P. Razaghi, F. Liu & L. Cai, Sporocadus cavernicola P. Razaghi, F. Liu & L. Cai, Sporocadus hyperici P. Razaghi, F. Liu & L. Cai, Sporocadus tibetensis P. Razaghi, F. Liu & L. Cai. New combinations: Discosia kaki (Kaz. Tanaka et al.) P. Razaghi, F. Liu & L. Cai, Monochaetia bulbophylli (S.F. Ran & Yong Wang bis) P. Razaghi, F. Liu & L. Cai, Neopestalotiopsis keteleeriae (Y. Song et al.) P. Razaghi, F. Liu & L. Cai. Citation: Razaghi P, Raza M, Han SL, Ma ZY, Cai L, Zhao P, Chen Q, Phurbu D, Liu F (2024). Sporocadaceae revisited. Studies in Mycology 109: 155-272. doi: 10.3114/sim.2024.109.03.
Keywords: Fungal diversity; Neopestalotiopsis; Pestalotiopsis; new taxa; plant pathogens; taxonomy.
© 2024 Westerdijk Fungal Biodiversity Institute.
Conflict of interest statement
The authors declare that there is no conflict of interest.
Figures



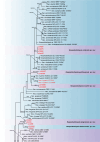



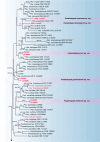





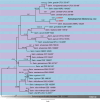

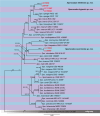























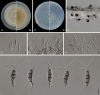

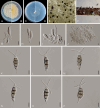







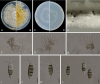





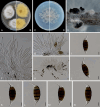




References
-
- Akinsanmi OA, Nisa S, Jeff-Ego OS. et al. (2017). Dry flower disease of Macadamia in Australia caused by Neopestalotiopsis macadamiae sp. nov. and Pestalotiopsis macadamiae sp. nov. Plant Disease 101: 45–53. - PubMed
-
- Andrews S, Babraham B. (2010). FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc
-
- Ariyawansa HA, Hyde KD. (2018). Additions to Pestalotiopsis in Taiwan. Mycosphere 9: 999–1013.
LinkOut - more resources
Full Text Sources
Miscellaneous
