Unraveling the genomic landscape of piscine myocarditis virus: mutation frequencies, viral diversity and evolutionary dynamics in Atlantic salmon
- PMID: 39717704
- PMCID: PMC11665822
- DOI: 10.1093/ve/veae097
Unraveling the genomic landscape of piscine myocarditis virus: mutation frequencies, viral diversity and evolutionary dynamics in Atlantic salmon
Abstract
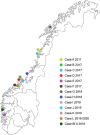
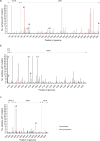
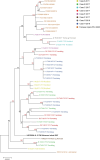
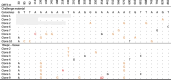
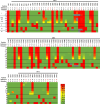
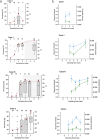
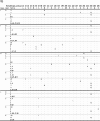
Over a decade since its discovery, piscine myocarditis virus (PMCV) remains a significant pathogen in Atlantic salmon aquaculture. Despite this significant impact, the genomic landscape, evolutionary dynamics, and virulence factors of PMCV are poorly understood. This study enhances the existing PMCV sequence dataset by adding 34 genome sequences and 202 new ORF3 sequences from clinical cardiomyopathy syndrome (CMS) cases in Norwegian aquaculture. Phylogenetic analyses, also including sequences from the Faroe Islands and Ireland revealed that PMCV sequences are highly conserved with distinct clustering by country of origin. Still, single CMS outbreaks display multiple PMCV variants, and although some clustering was seen by case origin, occasional grouping of sequences from different cases was also apparent. Temporal data from selected cases indicated increased sequence diversity in the population. We hypothesize that multiple bottlenecks and changing infection dynamics in the host population, with transfer to naïve individuals over time, represent a continuous selection pressure on the virus populations. No clear relation was found between PMCV variants and the severity of heart pathology. However, specific non-synonymous and synonymous mutations that might impact protein function and gene expression efficiency were identified. An additional factor that may impact PMCV replication is the presence of defective viral genomes, a novel finding for viruses of the order Ghabrivirales. This study provides new insights into PMCV genomic characteristics and evolutionary dynamics, highlighting the complex interplay of genetic diversity, virulence markers, and host-pathogen interactions, underscoring the epidemiological complexity of the virus. Keywords: piscine myocarditis virus; evolutionary dynamics; diversity; phylogeny; genomic sequencing; defective viral genomes.
Keywords: defective viral genomes; diversity; genomic sequencing; phylogeny; piscine myocarditis virus.
© The Author(s) 2024. Published by Oxford University Press.
Conflict of interest statement
M.L. was affiliated by the diagnostic service company PatoGen AS, which offers PMCV screening, when the analyses were performed. F.M. is affiliated by the aquaculture production company Mowi ASA.
Figures




Similar articles
-
Genomic analysis reveals low genetic diversity and no continuous reintroduction of piscine myocarditis virus in farmed Atlantic salmon in the Faroe Islands.J Gen Virol. 2025 Feb;106(2):002068. doi: 10.1099/jgv.0.002068. J Gen Virol. 2025. PMID: 39937589 Free PMC article.
-
Genetic diversity of piscine myocarditis virus in Atlantic salmon Salmo salar L. in Ireland.J Fish Dis. 2019 Aug;42(8):1161-1168. doi: 10.1111/jfd.13018. Epub 2019 Jun 6. J Fish Dis. 2019. PMID: 31169311
-
Comparing Genome Sequencing Methods to Reconstruct the Spread of Piscine Myocarditis Virus in Ireland.J Fish Dis. 2025 May 26:e14150. doi: 10.1111/jfd.14150. Online ahead of print. J Fish Dis. 2025. PMID: 40420515
-
Cardiomyopathy syndrome in Atlantic salmon Salmo salar L.: A review of the current state of knowledge.J Fish Dis. 2018 Jan;41(1):11-26. doi: 10.1111/jfd.12735. Epub 2017 Oct 24. J Fish Dis. 2018. PMID: 29064107 Review.
-
Antiviral defense in salmonids - Mission made possible?Fish Shellfish Immunol. 2019 Apr;87:421-437. doi: 10.1016/j.fsi.2019.01.043. Epub 2019 Jan 30. Fish Shellfish Immunol. 2019. PMID: 30708056 Review.
Cited by
-
Phylogenetic Diversity and Geographic Distribution of Atlantic Salmon Calicivirus in Major Salmon Farming Regions.J Fish Dis. 2025 Jun;48(6):e14107. doi: 10.1111/jfd.14107. Epub 2025 Feb 19. J Fish Dis. 2025. PMID: 39969098 Free PMC article.
-
Genomic analysis reveals low genetic diversity and no continuous reintroduction of piscine myocarditis virus in farmed Atlantic salmon in the Faroe Islands.J Gen Virol. 2025 Feb;106(2):002068. doi: 10.1099/jgv.0.002068. J Gen Virol. 2025. PMID: 39937589 Free PMC article.
-
Analysis of Piscine orthoreovirus genotype 1 genomes collected over a 32-year period (1988-2020) suggests a fitness peak in salmon hosts with minimal evidence for temporal divergence.Virus Evol. 2025 Jul 3;11(1):veaf033. doi: 10.1093/ve/veaf033. eCollection 2025. Virus Evol. 2025. PMID: 40630922 Free PMC article.
References
-
- Amin AB, Trasti J. Endomyocarditis in Atlantic salmon in Norwegian seafarms. Bull Eur Assoc Fish Pathol 1988;8:70–71.
-
- Bancroft JD, Stephens A. Theory and Practice of Histological Techniques. London: Churchill Livingstone, 1990.
LinkOut - more resources
Full Text Sources

