ZmNF-YB10, a maize NF-Y transcription factor, positively regulates drought and salt stress response in Arabidopsis thaliana
- PMID: 39718137
- PMCID: PMC11702966
- DOI: 10.1080/21645698.2024.2438421
ZmNF-YB10, a maize NF-Y transcription factor, positively regulates drought and salt stress response in Arabidopsis thaliana
Abstract
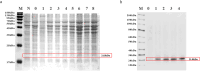
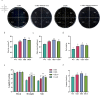
Maize (Zea mays L.) is a major food and feed crop and an important raw material for energy, chemicals, and livestock. The NF-Y family of transcription factors in maize plays a crucial role in the regulation of plant development and response to environmental stress. In this study, we successfully cloned and characterized the maize NF-Y transcription factor gene ZmNF-YB10. We used bioinformatics, quantitative fluorescence PCR, and other techniques to analyze the basic properties of the gene, its tissue expression specificity, and its role in response to drought, salt, and other stresses. The results indicated that the gene was 1209 base pairs (bp) in length, with a coding sequence (CDS) region of 618 bp, encoding a polypeptide composed of 205 amino acid residues. This polypeptide has a theoretical isoelectric point of 5.85 and features a conserved structural domain unique to the NF-Y family. Quantitative fluorescence PCR results demonstrated that the ZmNF-YB10 gene was differentially upregulated under drought and salt stress treatments but exhibited a negatively regulated expression pattern under alkali and cold stress treatments. Transgenic Arabidopsis thaliana subjected to drought and salt stress in soil showed greener leaves than wild-type A. thaliana. In addition, the overexpression lines showed reduced levels of hydrogen peroxide (H2O2), superoxide (O2-), and malondialdehyde (MDA) and increased activities of peroxidase (POD), catalase (CAT), and superoxide dismutase (SOD). Western blot analysis revealed a distinct band at 21.8 kDa. Salt and drought tolerance analyses conducted in E. coli BL21 indicated a positive regulation. In yeast cells, ZmNF-YB10 exhibited a biological function that enhances salt and drought tolerance. Protein interactions were observed among the ZmNF-YB10, ZmNF-YC2, and ZmNF-YC4 genes. It is hypothesized that the ZmNF-YB10, ZmNF-YC2, and ZmNF-YC4 genes may play a role in the response to abiotic stresses, such as drought and salt tolerance, in maize.
Keywords: Arabidopsis; drought stress; maize; salt stress; yeast heterologous expression.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures
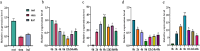






Similar articles
-
Overexpression of maize transcription factor ZmNF-YC14 positively regulates drought and salt stress responses in Arabidopsis thaliana.Plant Sci. 2025 Jul;356:112502. doi: 10.1016/j.plantsci.2025.112502. Epub 2025 Apr 7. Plant Sci. 2025. PMID: 40204192
-
Isolation, structural analysis, and expression characteristics of the maize nuclear factor Y gene families.Biochem Biophys Res Commun. 2016 Sep 16;478(2):752-8. doi: 10.1016/j.bbrc.2016.08.020. Epub 2016 Aug 4. Biochem Biophys Res Commun. 2016. PMID: 27498027
-
Transcription factors ZmNF-YA1 and ZmNF-YB16 regulate plant growth and drought tolerance in maize.Plant Physiol. 2022 Sep 28;190(2):1506-1525. doi: 10.1093/plphys/kiac340. Plant Physiol. 2022. PMID: 35861438 Free PMC article.
-
Drought stress memory in maize: understanding and harnessing the past for future resilience.Plant Cell Rep. 2025 Apr 25;44(5):101. doi: 10.1007/s00299-025-03494-x. Plant Cell Rep. 2025. PMID: 40278890 Review.
-
Interplay between auxin and abiotic stresses in maize.J Exp Bot. 2025 May 10;76(7):1879-1887. doi: 10.1093/jxb/eraf079. J Exp Bot. 2025. PMID: 40159933 Free PMC article. Review.
References
-
- Nelson DE, Repetti PP, Adams TR, Creelman RA, Wu J, Warner DC, Anstrom DC, Bensen RJ, Castiglioni PP, Donnarummo MG, et al. Plant nuclear factor Y (NF-Y) B subunits confer drought tolerance and lead to improved corn yields on water-limited acres. Proc Natl Acad Sci USA. 2007;104(42):16450–55. doi:10.1073/pnas.0707193104. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
