Microarray-Based Methodology for Lipid Profiling, Enzymatic Activity, And Binding Assays in Printed Lipid Raft Membranes from Astrocytes and Neurons
- PMID: 39718364
- PMCID: PMC11740170
- DOI: 10.1021/acs.analchem.4c02421
Microarray-Based Methodology for Lipid Profiling, Enzymatic Activity, And Binding Assays in Printed Lipid Raft Membranes from Astrocytes and Neurons
Abstract
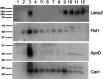
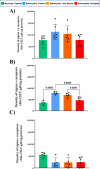
Lipid rafts are liquid-ordered domains in which specific enzymes and receptors are located. These membrane platforms play crucial roles in a variety of signaling pathways. Alterations in the lipid environment, such as those elicited by oxidative stress, can lead to important functional disruptions in membrane proteins. Cell membrane microarrays have emerged in the past decade as a powerful methodology for the study of both lipids and membrane proteins at large scales. Based on that technology and the importance of liquid-ordered subdomains, we have developed a new printed lipid raft technology with a preserved native protein structure and lipid environment. To validate this technology and evaluate its potential for different aims, raft membrane microarrays (RMMAs) containing two different cell types (astrocytes and neurons) and three different conditions (astrocytes in control situation, metabolic stress, and oxidative stress) were developed. To study differences in lipid profiles between raft domains, the MALDI-MS assay was performed on RMMAs. To evaluate the preservation of native protein activities (enzymatic activity and ligand binding) in the printed raft domains, differences in NADH oxidoreductase, GAPDH, cholinesterase activities, and sigma-1 and sigma-2 binding assays were performed. We demonstrate the performance of this new microarray technology, adapted to membrane subdomains, as valid to explore changes in lipid composition and protein activities in raft domains from brain cell lines under different stress conditions relevant for neuropathology.
Conflict of interest statement
The authors declare no competing financial interest.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

