Transcriptome Profiling, Cloning, and Characterization of AnGlu04478, a Ginsenoside Hydrolyzing β-Glucosidase from Aspergillus niger NG1306
- PMID: 39718650
- PMCID: PMC11668888
- DOI: 10.1007/s00284-024-04012-0
Transcriptome Profiling, Cloning, and Characterization of AnGlu04478, a Ginsenoside Hydrolyzing β-Glucosidase from Aspergillus niger NG1306
Abstract
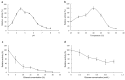
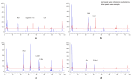
β-Glucosidase plays a pivotal role in transforming ginsenosides into specific minor ginsenosides. In this study, total ginsenosides from Panax notoginseng leaves were used as substrates to stimulate the growth of Aspergillus niger NG1306. Transcriptome analysis identified a β-glucosidase gene, Anglu04478 (1455 bp, 484 amino acids, 54.5 kDa, pI = 5.1), as a participant in the ginsenosides biotransformation process. This gene was cloned and expressed in Escherichia coli BL21 Transetta (DE3). The AnGlu04478 protein was purified using a Ni2+ column, and its enzymatic properties were characterized. Purified AnGlu04478 exhibited a specific activity of 32.97 U/mg when assayed against pNPG. Under optimal conditions (pH 4.5, temperature 40 °C), the kinetic parameters, Km and Vmax, for pNPG were 1.55 mmol/L and 0.014 mmol/min, respectively. Cu2+ displayed an inhibitory effect on AnGlu04478, whereas Ca2+, Co2+, and Ni2+ ions had minimal impact. The enzyme showed tolerance to ethanol and was largely unaffected by glucose feedback inhibition. Testing with ginsenosides as substrates revealed selective hydrolysis at the C3 position of ginsenosides Rb1, Rb2, Rb3, and Rc, with the metabolic pathway delineated as Rb1 → GypXVII, Rb2 → C-O, Rb3 → C-Mx1 → C-Mx, and Rc → C-Mc1. The conversion rates of ginsenosides Rb1, Rb2, Rb3, and Rc varied from 2.58 to 20.63%. With 0.5 U purified enzyme and 0.5 mg total ginsenosides, incubated at 40 °C for 12 h, the conversion rates were 42.6% for GypXVII, 10.4% for C-O, 6.27% for C-Mx1, 26.96% for C-Mx, and 90% for Rc. These results suggest that AnGlu04478 displays substrate promiscuity as a β-glucosidase, thus broadening the potential for ginsenoside biotransformation.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Conflict of interest: The authors have no financial conflicts of interest to declare.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

