New estimates and synthesis of chromosome numbers, ploidy levels and genome size variation in Allium sect. Codonoprasum: advancing our understanding of the unresolved diversification and evolution of this section
- PMID: 39718713
- PMCID: PMC11668721
- DOI: 10.1186/s40529-024-00446-8
New estimates and synthesis of chromosome numbers, ploidy levels and genome size variation in Allium sect. Codonoprasum: advancing our understanding of the unresolved diversification and evolution of this section
Abstract
Background: The genus Allium is known for its high chromosomal variability, but most chromosome counts are based on a few individuals and genome size (GS) reports are limited in certain taxonomic groups. This is evident in the Allium sect. Codonoprasum, a species-rich (> 150 species) and taxonomically complex section with weak morphological differences between taxa, the presence of polyploidy and frequent misidentification of taxa. Consequently, a significant proportion of older karyological reports may be unreliable and GS data are lacking for the majority of species within the section. This study, using chromosome counting and flow cytometry (FCM), provides the first comprehensive and detailed insight into variation in chromosome number, polyploid frequency and distribution, and GS in section members, marking a step towards understanding the unresolved diversification and evolution of this group.
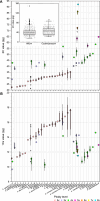
Results: We analysed 1578 individuals from 316 populations of 25 taxa and reported DNA ploidy levels and their GS, with calibration from chromosome counts in 22 taxa. Five taxa had multiple ploidy levels. First estimates of GS were obtained for 16 taxa. A comprehensive review of chromosome number and DNA-ploidy levels in 129 taxa of the section revealed that all taxa have x = 8, except A. rupestre with two polyploid series (x = 8, descending dysploidy x = 7), unique for this section. Diploid taxa dominated (72.1%), while di- & polyploid (12.4%) and exclusively polyploid (15.5%) taxa were less common. Ploidy diversity showed that diploid taxa dominated in the eastern Mediterranean and decreased towards the west and north, whereas only polyploid cytotypes of di- & polyploid taxa or exclusively polyploid taxa dominated in northern and northwestern Europe. A 4.1-fold variation in GS was observed across 33 taxa analysed so far (2C = 22.3-92.1 pg), mainly due to polyploidy, with GS downsizing observed in taxa with multiple ploidy levels. Intra-sectional GS variation suggests evolutionary relationships, and intraspecific GS variation within some taxa may indicate taxonomic heterogeneity and/or historical migration patterns.
Conclusions: Our study showed advantages of FCM as an effective tool for detecting ploidy levels and determining GS within the section. GS could be an additional character in understanding evolution and phylogenetic relationships within the section.
Keywords: Chromosome number; Cytogeography; DNA ploidy level; Flow cytometry; Genome size; Polyploidy.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare that they have no competing interests.
Figures


Similar articles
-
Cytogeography of the Humifusa clade of Opuntia s.s. Mill. 1754 (Cactaceae, Opuntioideae, Opuntieae): correlations with pleistocene refugia and morphological traits in a polyploid complex.Comp Cytogenet. 2012 Feb 14;6(1):53-77. doi: 10.3897/CompCytogen.v6i1.2523. eCollection 2012. Comp Cytogenet. 2012. PMID: 24260652 Free PMC article.
-
Genome Size and Chromosome Number Evaluation of Astragalus L. sect. Hymenostegis Bunge (Fabaceae).Plants (Basel). 2022 Feb 5;11(3):435. doi: 10.3390/plants11030435. Plants (Basel). 2022. PMID: 35161416 Free PMC article.
-
Role of adaptive and non-adaptive mechanisms forming complex patterns of genome size variation in six cytotypes of polyploid Allium oleraceum (Amaryllidaceae) on a continental scale.Ann Bot. 2013 Mar;111(3):419-31. doi: 10.1093/aob/mcs297. Epub 2013 Jan 24. Ann Bot. 2013. PMID: 23348752 Free PMC article.
-
Allium cytogenetics: a critical review on the Indian taxa.Comp Cytogenet. 2023 May 29;17:129-156. doi: 10.3897/CompCytogen.17.98903. eCollection 2023. Comp Cytogenet. 2023. PMID: 37304149 Free PMC article. Review.
-
Preliminary Review of the Diploid Taxa in Hieracium s.s.Plants (Basel). 2025 Mar 29;14(7):1057. doi: 10.3390/plants14071057. Plants (Basel). 2025. PMID: 40219125 Free PMC article. Review.
References
-
- Aedo C (2013) Allium L. In: Rico E, Crespo MB, Quintanar A, Herrero A, Aedo C (eds) Flora Iberica, Liliaceae-Agavaceae, vol 20. Editorial CSIC, Madrid, pp 220–273
-
- Afonso A, Loureiro J, Arroyo J, Olmedo-Vicente E, Castro S (2021) Cytogenetic diversity in the polyploid complex Linum suffruticosum s.l. (Linaceae). Bot J Linn Soc 195:216–232. 10.1093/botlinnean/boaa060
-
- Alden B (1976) Floristic reports from the high mountains of Pindhos, Greece. Bot Not 129:297–321
-
- Anačkov G (2009) Taxonomy and chorology of genus Allium L. 1754 (Amaryllidales, Alliaceae) in Serbia. Dissertation, University of Novi Sad, Serbia
-
- Andersson IA (1991) Allium L. In: Strid A, Tan K (eds) Mountain Flora of Greece, 2. Edinburgh University Press, Edinburgh, pp 701–714
Grants and funding
LinkOut - more resources
Full Text Sources

