Targeted degradation of oncogenic KRASG12V triggers antitumor immunity in lung cancer models
- PMID: 39718828
- PMCID: PMC11735103
- DOI: 10.1172/JCI174249
Targeted degradation of oncogenic KRASG12V triggers antitumor immunity in lung cancer models
Abstract
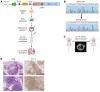
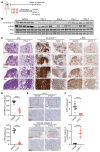
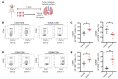
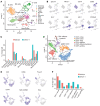
Kirsten rat sarcoma viral oncogene homolog (KRAS) is the most frequently mutated oncogene in lung adenocarcinoma, with G12C and G12V being the most predominant forms. Recent breakthroughs in KRASG12C inhibitors have transformed the clinical management of patients with the G12C mutation and advanced our understanding of the function of this mutation. However, little is known about the targeted disruption of KRASG12V, partly due to a lack of specific inhibitors. Here, we leverage the degradation tag (dTAG) system to develop a KRASG12V-transgenic mouse model. We explored the therapeutic potential of KRASG12V degradation and characterized its effect on the tumor microenvironment (TME). Our study reveals that degradation of KRASG12V abolished lung and pancreatic tumors in mice and caused a robust inhibition of KRAS-regulated cancer-intrinsic signaling. Importantly, targeted degradation of KRASG12V reprogrammed the TME toward a stimulatory milieu and drove antitumor immunity, elicited mainly by effector and cytotoxic CD8+ T cells. Our work provides insights into the effect of KRASG12V degradation on both tumor progression and the immune response, highlighting degraders as a powerful strategy for targeting KRAS-mutant cancers.
Keywords: Immunology; Lung cancer; Oncology.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

