A conserved human CD4+ T cell subset recognizing the mycobacterial adjuvant trehalose monomycolate
- PMID: 39718834
- PMCID: PMC11910211
- DOI: 10.1172/JCI185443
A conserved human CD4+ T cell subset recognizing the mycobacterial adjuvant trehalose monomycolate
Abstract
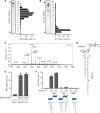
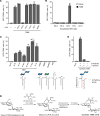
Mycobacterium tuberculosis causes human tuberculosis (TB). As mycobacteria are protected by a thick lipid cell wall, humans have developed immune responses against diverse mycobacterial lipids. Most of these immunostimulatory lipids are known as adjuvants acting through innate immune receptors, such as C-type lectin receptors. Although a few mycobacterial lipid antigens activate unconventional T cells, the antigenicity of most adjuvantic lipids is unknown. Here, we identified that trehalose monomycolate (TMM), an abundant mycobacterial adjuvant, activated human T cells bearing a unique αβ T cell receptor (αβTCR). This recognition was restricted by CD1b, a monomorphic antigen-presenting molecule conserved in primates but not mice. Single-cell TCR-RNA-Seq using newly established CD1b-TMM tetramers revealed that TMM-specific T cells were present as CD4+ effector memory T cells in the periphery of uninfected donors but expressed IFN-γ, TNF, and anti-mycobacterial effectors upon TMM stimulation. TMM-specific T cells were detected in cord blood and PBMCs of donors without bacillus Calmette-Guérin vaccination but were expanded in patients with active TB. A cryo-electron microscopy study of CD1b-TMM-TCR complexes revealed unique antigen recognition by conserved features of TCRs, positively charged CDR3α, and long CDR3β regions. These results indicate that humans have a commonly shared and preformed CD4+ T cell subset recognizing a typical mycobacterial adjuvant as an antigen. Furthermore, the dual role of TMM justifies reconsideration of the mechanism of action of adjuvants.
Keywords: Immunology; Infectious disease; Structural biology; T cell receptor; Tuberculosis.
Figures





Comment in
- Sensing mycobacteria through unconventional pathways doi: 10.1172/JCI190230
References
-
- Freund J, et al. Sensitization and antibody formation after injection of tubercle bacilli and paraffin oil. Proc Soc Exp Biol Med. 1937;37(3):509. doi: 10.3181/00379727-37-9625. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

