Distinct transcriptional changes distinguish efficient and poor remyelination in multiple sclerosis
- PMID: 39718981
- PMCID: PMC12129743
- DOI: 10.1093/brain/awae414
Distinct transcriptional changes distinguish efficient and poor remyelination in multiple sclerosis
Abstract
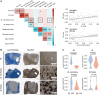
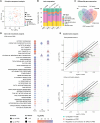
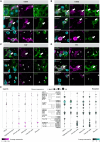
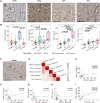
Multiple sclerosis (MS) is a highly heterogeneous disease, with varying remyelination potential across individuals and between lesions. However, the molecular mechanisms underlying the potential to remyelinate remain poorly understood. In this study, we aimed to take advantage of the intrinsic heterogeneity in remyelinating capacity between MS donors and lesions to uncover known and novel pro-remyelinating molecules for MS therapies. To elucidate distinct molecular signatures underlying the potential to remyelinate, we stratified MS donors from the Netherlands Brain Bank cohort (n = 239), based on proportions of remyelinated lesions (RLs), into efficiently remyelinating donors (ERDs; n = 21) and poorly remyelinating donors (PRDs; n = 19). We performed bulk RNA sequencing of RLs, active lesions with ramified and amoeboid microglia/macrophage morphology (ALs non-foamy), active lesions with foamy microglia/macrophage morphology (ALs foamy) and normal-appearing white matter (NAWM) from ERDs and PRDs. We found that ALs non-foamy were positively correlated with remyelination, whereas ALs foamy were not, indicating a role for microglia/macrophage state in influencing remyelination potential. Bioinformatics analyses were performed to identify key pathways and molecules implicated in the remyelination process. We found distinct differences between the donors with differing remyelination potential in comparable MS lesion types. The RLs and ALs non-foamy of ERDs versus PRDs showed upregulation of the epithelial-mesenchymal transition pathway, whereas in ALs foamy of PRDs, inflammation and damage-associated pathways (i.e. MTORC1 signalling, TNF signalling and oxidative phosphorylation) were upregulated in comparison to ALs foamy of ERDs, suggesting that these latter pathways might counteract remyelination. We found genes significantly upregulated in RLs and/or ALs non-foamy of ERDs that have previously been associated with remyelination, including CXCL12, EGF, HGF, IGF2, IL10, PDGFB, PPARG and TREM2, illustrating the strength of our donor and lesion stratification. TGFB1, TGFB2, EGF and IGF1 were determined to be key upstream regulators of genes upregulated in RLs and ALs non-foamy of ERDs. We also identified potential novel pro-remyelinating molecules, such as BTC, GDF10, GDF15, CCN1, CCN4, FGF5, FGF10 and INHBB. Our study identified both known and novel genes associated with efficient remyelination that might facilitate the development of therapeutic strategies to promote tissue repair and clinical recovery in MS.
Keywords: active lesions; growth factors; microglia; repair.
© The Author(s) 2024. Published by Oxford University Press on behalf of the Guarantors of Brain.
Conflict of interest statement
The authors report no competing interests.
Figures

References
-
- Lassmann H, Raine CS, Antel J, Prineas JW. Immunopathology of multiple sclerosis: Report on an international meeting held at the Institute of Neurology of the University of Vienna. J Neuroimmunol. 1998;86:213–217. - PubMed
-
- Lucchinetti C, Brück W, Parisi J, Scheithauer B, Rodriguez M, Lassmann H. Heterogeneity of multiple sclerosis lesions: Implications for the pathogenesis of demyelination. Ann Neurol. 2000;47:707–717. - PubMed
-
- Franklin RJM, Ffrench-Constant C. Regenerating CNS myelin—From mechanisms to experimental medicines. Nat Rev Neurosci. 2017;18:753–769. - PubMed
-
- Franklin RJM, Simons M. CNS remyelination and inflammation: From basic mechanisms to therapeutic opportunities. Neuron. 2022;110:3549–3565. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

