A subcellular selective APEX2-based proximity labeling used for identifying mitochondrial G-quadruplex DNA binding proteins
- PMID: 39718986
- PMCID: PMC11724306
- DOI: 10.1093/nar/gkae1259
A subcellular selective APEX2-based proximity labeling used for identifying mitochondrial G-quadruplex DNA binding proteins
Abstract
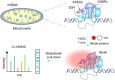
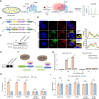
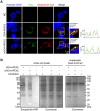
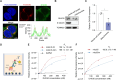
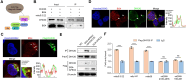
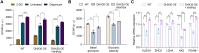
G-quadruplexes (G4s), as an important type of non-canonical nucleic acid structure, have received much attention because of their regulations of various biological processes in cells. Identifying G4s-protein interactions is essential for understanding G4s-related biology. However, current strategies for exploring G4 binding proteins (G4BPs) include pull-down assays in cell lysates or photoaffinity labeling, which are lack of sufficient spatial specificity at the subcellular level. Herein, we develop a subcellular selective APEX2-based proximity labeling strategy to investigate the interactome of mitochondrial DNA (mtDNA) G4s in living cells. By this method, we have identified several mtDNA G4BPs. Among them, a previously unrecognized mtDNA G4BP, DHX30 has been selected as an example to explore its important biofunctions. DHX30 localizes both in cytoplasm and mitochondria and can resolve mtDNA G4s. Further studies have demonstrated that DHX30 unfolds mtDNA G4 in living cells, which results in a decrease in glycolysis activity of tumor cells. Besides, RHPS4, a known mtDNA G4 stabilizer, will reverse this inhibition effect. Benefiting from the high spatiotemporal resolution and the ability of genetically encoded systems to perform the labeling with exquisite specificity within living cells, our approach can realize the identification of subcellular localized G4BPs. Our work provides a novel strategy to map protein interactions of specific nucleic acid features in subcellular compartments of living cells.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

