Post-eclosion growth in the Drosophila ejaculatory duct is driven by Juvenile hormone signaling and is essential for male fertility
- PMID: 39719194
- PMCID: PMC12051359
- DOI: 10.1016/j.ydbio.2024.12.010
Post-eclosion growth in the Drosophila ejaculatory duct is driven by Juvenile hormone signaling and is essential for male fertility
Abstract
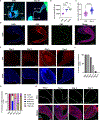
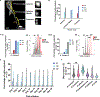
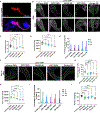
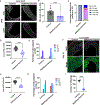
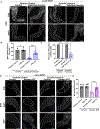
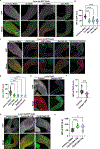
The Drosophila Ejaculatory duct (ED) is a secretory tissue of the somatic male reproductive system. The ED is involved in the secretion of seminal fluid components and ED-specific antimicrobial peptides that aid in fertility and the female post-mating response. The ED is composed of secretory epithelial cells surrounded by a layer of innervated contractile muscle. The ED grows in young adult males during the first 24 h post-eclosion, but the cell cycle status of the ED secretory cells and the role of post-eclosion ED growth have been unexplored. Here, we show that secretory cells of the adult Drosophila ED undergo variant cell cycles lacking mitosis called the endocycle, that lead to an increase in the cell and organ size of the ED post eclosion. The cells largely exit the endocycle by day 3 of adulthood, when the growth of the ED ceases, resulting in a tissue containing cells of ploidies ranging from 8C to 32C. The size of the ED directly correlates with the ploidy of the secretory cells, with additional ectopic endocycles increasing organ size. When endoreplication is compromised in ED secretory cells, it leads to reduced organ size, reduced protein synthesis and compromised fertility. We provide evidence that the growth and endocycling in the young adult male ED is dependent on Juvenile hormone (JH) signaling and we suggest that hormone-induced early adult endocycling is required for optimal fertility and function of the ED tissue. We propose to use the ED as a post-mitotic tissue model to study the role of polyploidy in regulating secretory tissue growth and function.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Figures





Update of
-
Post-eclosion growth in the Drosophila Ejaculatory Duct is driven by Juvenile Hormone signaling and is essential for male fertility.bioRxiv [Preprint]. 2024 Aug 13:2024.08.12.607650. doi: 10.1101/2024.08.12.607650. bioRxiv. 2024. Update in: Dev Biol. 2025 Mar;519:122-141. doi: 10.1016/j.ydbio.2024.12.010. PMID: 39185157 Free PMC article. Updated. Preprint.
Similar articles
-
Post-eclosion growth in the Drosophila Ejaculatory Duct is driven by Juvenile Hormone signaling and is essential for male fertility.bioRxiv [Preprint]. 2024 Aug 13:2024.08.12.607650. doi: 10.1101/2024.08.12.607650. bioRxiv. 2024. Update in: Dev Biol. 2025 Mar;519:122-141. doi: 10.1016/j.ydbio.2024.12.010. PMID: 39185157 Free PMC article. Updated. Preprint.
-
Cell cycle variants during Drosophila male accessory gland development.G3 (Bethesda). 2024 Jul 8;14(7):jkae089. doi: 10.1093/g3journal/jkae089. G3 (Bethesda). 2024. PMID: 38683731 Free PMC article.
-
Control of Drosophila endocycles by E2F and CRL4(CDT2).Nature. 2011 Oct 30;480(7375):123-7. doi: 10.1038/nature10579. Nature. 2011. PMID: 22037307 Free PMC article.
-
Developmental control of endocycles and cell growth in plants.Curr Opin Plant Biol. 2010 Dec;13(6):654-60. doi: 10.1016/j.pbi.2010.10.006. Epub 2010 Nov 18. Curr Opin Plant Biol. 2010. PMID: 21094078 Review.
-
Endocycles: a recurrent evolutionary innovation for post-mitotic cell growth.Nat Rev Mol Cell Biol. 2014 Mar;15(3):197-210. doi: 10.1038/nrm3756. Nat Rev Mol Cell Biol. 2014. PMID: 24556841 Review.
Cited by
-
A window of cell cycle plasticity enables imperfect regeneration of an adult postmitotic organ in Drosophila.bioRxiv [Preprint]. 2025 Jun 6:2025.06.05.658044. doi: 10.1101/2025.06.05.658044. bioRxiv. 2025. PMID: 40501567 Free PMC article. Preprint.
References
-
- Ahmad SM, Baker BS, 2002. Sex-specific deployment of FGF signaling in Drosophila recruits mesodermal cells into the male genital imaginal disc. Cell 109, 651–661. - PubMed
-
- Avila FW, Sánchez-López JA, McGlaughon JL, Raman S, Wolfner MF, Heifetz Y, 2016. Nature and Functions of Glands and Ducts in the Drosophila Reproductive Tract, in: Cohen E, Moussian B (Eds.), Extracellular Composite Matrices in Arthropods. Springer International Publishing, Cham, pp. 411–444.
-
- Bairati A, 1968. STRUCTURE AND ULTRASTRUCTURE OF THE MALE REPRODUCTIVE SYSTEM IN DROSOPHILA MELANOGASTER MEIG. Monitore Zoologico Italiano - Italian Journal of Zoology 2:3–4, 105–182.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

