Structure and dynamics of the interaction of Delta and Omicron BA.1 SARS-CoV-2 variants with REGN10987 Fab reveal mechanism of antibody action
- PMID: 39719448
- PMCID: PMC11668877
- DOI: 10.1038/s42003-024-07422-9
Structure and dynamics of the interaction of Delta and Omicron BA.1 SARS-CoV-2 variants with REGN10987 Fab reveal mechanism of antibody action
Abstract
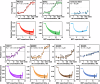
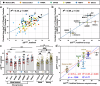
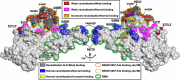
Study of mechanisms by which antibodies recognize different viral strains is necessary for the development of new drugs and vaccines to treat COVID-19 and other infections. Here, we report 2.5 Å cryo-EM structure of the SARS-CoV-2 Delta trimeric S-protein in complex with Fab of the recombinant analog of REGN10987 neutralizing antibody. S-protein adopts "two RBD-down and one RBD-up" conformation. Fab interacts with RBDs in both conformations, blocking the recognition of angiotensin converting enzyme-2. Three-dimensional variability analysis reveals high mobility of the RBD/Fab regions. Interaction of REGN10987 with Wuhan, Delta, Omicron BA.1, and mutated variants of RBDs is analyzed by microscale thermophoresis, molecular dynamics simulations, and ΔG calculations with umbrella sampling and one-dimensional potential of mean force. Variability in molecular dynamics trajectories results in a large scatter of calculated ΔG values, but Boltzmann weighting provides an acceptable correlation with experiment. REGN10987 evasion of the Omicron variant is found to be due to the additive effect of the N440K and G446S mutations located at the RBD/Fab binding interface with a small effect of Q498R mutation. Our study explains the influence of known-to-date SARS-CoV-2 RBD mutations on REGN10987 recognition and highlights the importance of dynamics data beyond the static structure of the RBD/Fab complex.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures






References
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

