Evolution and comparative transcriptome analysis of glucosinolate pathway genes in Brassica napus L
- PMID: 39719940
- PMCID: PMC11666375
- DOI: 10.3389/fpls.2024.1483635
Evolution and comparative transcriptome analysis of glucosinolate pathway genes in Brassica napus L
Abstract
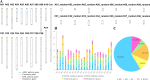
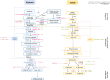
Glucosinolates (GSLs) are important secondary metabolites abundantly distributed in Brassicaceae plants, whose degradation products benefit plant resistance but are regarded as disadvantageous factors for human health. Thus, reducing GSL content is an important goal in the breeding program in crops, such as Brassica napus. In this study, 1280 genes in the GSL pathway were identified from 14 land plant genomes, which are specifically distributed in Brassicaceae and are extensively expanded in B. napus. Most GSL pathway genes had many positive selection sites, especially the encoding genes of transcription factors (TFs) and structural genes involved in the GSL breakdown process. There are 344 genes in the GSL pathway in the B. napus genome, which are unequally distributed on the 19 chromosomes. Whole-genome duplication mainly contributed to the gene expansion of the GSL pathway in B. napus. The genes in GSL biosynthesis were regulated by various TFs and cis-elements in B. napus and mainly response to abiotic stress and hormone induction. A comparative transcriptome atlas of the roots, stems, leaves, flowers, siliques, and seeds of a high- (ZY821), and a low-GSL-content (ZS11) cultivar was constructed. The features of the two cultivars may be attributed to diverse expression differences in each organ at different stages, especially in seeds. In all, 65 differential expressed genes (DEGs) concentrated on the core structure pathway were inferred to mainly influence the GSL contents between ZY821 and ZS11. This study provides an important RNA-seq dataset and diverse gene resources for future manipulating GSLs biosynthesis and distribution in B. napus using molecular breeding methods.
Keywords: Brassica napus; evolution; glucosinolate; pathway; transcriptome.
Copyright © 2024 Liu, Wu, Chen, Chen, Shen, Qadir, Wan, Zhao, Yin, Li, Qu and Du.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
LinkOut - more resources
Full Text Sources

