Identification of potential molecular markers for detection of lengthy chilled storage of Prunus persica L. fruit
- PMID: 39720059
- PMCID: PMC11667614
- DOI: 10.1016/j.heliyon.2024.e40992
Identification of potential molecular markers for detection of lengthy chilled storage of Prunus persica L. fruit
Abstract
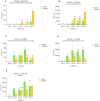
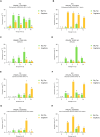
Low temperature is the main strategy to preserve fruit quality post-harvest, in the supply chain. Low temperatures reduce the respiration, ethylene emission, and enzymatic activities associated with senescence. Unfortunately, peaches are sensitive to low temperatures if exposed for long periods, resulting in physiological disorders that can compromise commercial quality. Maximum damage occurs at 5 °C while at 1 °C damage is reduced. Therefore, rapid early detection methods for the distribution chain to monitor length and temperature of fruit storage are needed. The aim of this work was to identify candidate genes to develop an antibody-based marker system in peach fruit to monitor chilled storage. Two cultivars were tested: 'Sagittaria', an early ripening peach, and 'Big Top' a mid-season ripening nectarine, with delayed softening and resistance to supply-chain conditions. Both cultivars were subjected to 1 or 5 °C chilled storage for different times to simulate typical supply-chain conditions. Identification and expression of potential marker genes was assessed using a previous transcriptomic study following storage at 1 °C. Fifteen candidate genes were selected, however only seven proteins encoded were suitable as protein markers as they lack a transmembrane domain. Real-time qPCR using fruit from the subsequent year to the transcriptome was used to assess expression at both 1 and 5 °C chilled storage of five candidate genes. Four genes and the related proteins were identified that would be suitable for the development of molecular markers: a Pathogenesis-Related Bet v I family protein, a dehydrin, a Glycosyl hydrase family 18 protein and a Late Embryogenesis abundant protein.
Keywords: Cold storage; ELISA test; Gene expression; Molecular markers; Prunus persica L.
© 2024 The Authors. Published by Elsevier Ltd.
Conflict of interest statement
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:Natasha Damiana Spadafora reports financial support was provided by CON IL SUD Foundation. Natasha Damiana Spadafora reports financial support was provided by PON. If there are other authors, they declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


References
-
- FAOSTAT Productions of peaches and nectarines in 2022. 2022. http://www.fao.org/faostat/en/#data/QC
-
- Farina V., Lo Bianco R., Mazzaglia A. Evaluation of late-maturing peach and nectarine fruit quality by chemical, physical, and sensory determinations. Agriculture. 2019;9:189. doi: 10.3390/agriculture9090189. - DOI
-
- Farooq S., Mir M.M., Ganai S.A., Maqbool T., Mir S.A., Shah M.A. In: Postharvest Biology and Technology of Temperate Fruits. Mir S., Shah M., Mir M., editors. Springer; Cham: 2018. Postharvest biology and technology of peach. - DOI
-
- Aubert C., Bony P., Chalot G., Landry P., Lurol S. Effects of storage temperature, storage duration, and subsequent ripening on the physicochemical characteristics, volatile compounds, and phytochemicals of western red nectarine (Prunus persica L. Batsch) J. Agric. Food Chem. 2014;62:4707–4724. doi: 10.1021/jf4057555. - DOI - PubMed
-
- Onwude D.I., Chen G., Eke-emezie N., Kabutey A., Khaled A.Y., Sturm B. Recent advances in reducing food losses in the supply chain of fresh agricultural produce. Processes. 2020;8:1431. doi: 10.3390/pr8111431. - DOI

