Comprehensive evaluation of AlphaMissense predictions by evidence quantification for variants of uncertain significance
- PMID: 39720176
- PMCID: PMC11666499
- DOI: 10.3389/fgene.2024.1487608
Comprehensive evaluation of AlphaMissense predictions by evidence quantification for variants of uncertain significance
Abstract
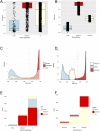
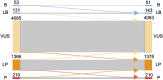
Accurate variant classification is critical for genetic diagnosis. Variants without clear classification, known as "variants of uncertain significance" (VUS), pose a significant diagnostic challenge. This study examines AlphaMissense performance in variant classification, specifically for VUS. A systematic comparison between AlphaMissense predictions and predictions based on curated evidence according to the ACMG/AMP classification guidelines was conducted for 5845 missense variants in 59 genes associated with representative Mendelian disorders. A framework for quantifying and modeling VUS pathogenicity was used to facilitate comparison. Manual reviewing classified 5845 variants as 4085 VUS, 1576 pathogenic/likely pathogenic, and 184 benign/likely benign. Pathogenicity predictions based on AlphaMissense and ACMG guidelines were concordant for 1887 variants (1352 pathogenic, 132 benign, and 403 VUS/ambiguous). The sensitivity and specificity of AlphaMissense predictions for pathogenicity were 92% and 78%. Moreover, the quantification of VUS evidence and heatmaps weakly correlated with the AlphaMissense score. For VUS without computational evidence, incorporating AlphaMissense changed the VUS quantification for 878 variants, while 56 were reclassified as likely pathogenic. When AlphaMissense replaced existing computational evidence for all VUS, 1709 variants changed quantified criteria while 63 were reclassified as likely pathogenic. Our research suggests that the augmentation of AlphaMissense with empirical evidence may improve performance by incorporating a quantitative framework to aid in VUS classification.
Keywords: ACMG/AMP classification; AlphaMissense; VUS; variant classification; variants of unknown significance.
Copyright © 2024 Kurtovic-Kozaric, Delalic, Mutapcic, Comor, Siciliano and Kiel.
Conflict of interest statement
AK-K, LD, BM, LC, ES, and MK were employed by the company Genomenon.
Figures


References
-
- Ahmad R. N. R., Zhang L. T., Morita R., Tani H., Wu Y., Chujo T., et al. (2024). Pathological mutations promote proteolysis of mitochondrial tRNA-specific 2-thiouridylase 1 (MTU1) via mitochondrial caseinolytic peptidase (CLPP). Nucleic Acids Res. 52 (3), 1341–1358. 10.1093/nar/gkad1197 - DOI - PMC - PubMed
-
- Barboso G., Fragnito C., Beghi C., Saccani S., Fesani F. (1987). Asymptomatic patient reoperated on for severe proximal stenosis of circular sequential vein graft. J. Cardiovasc Surg. 28 (3), 341–342. - PubMed
LinkOut - more resources
Full Text Sources

