Prediction of key biological processes from intercellular DNA damage differences through model-based fitting
- PMID: 39720517
- PMCID: PMC11667071
- DOI: 10.1016/j.isci.2024.111473
Prediction of key biological processes from intercellular DNA damage differences through model-based fitting
Abstract
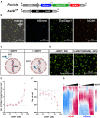
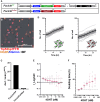
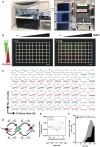
DNA double-strand breaks (DSBs) occurring within the genomic DNA of mammalian cells significantly impact cell survival, depending upon their repair capacity. This study presents a mathematical model to fit fibroblast survival rates with a sequence-specific DSB burden induced by the restriction enzyme AsiSI. When cells had a sporadic DSB burden under mixed culture, cell growth showed a good fit to the Lotka-Volterra competitive equation, predicting the presence of modifying factors acting as competitive cell-to-cell interactions compared to monocultures. Under the predicted condition, we found the Acta2 gene, a known marker of cancer-associated fibroblasts, played a role in competitive interactions between cells with different DSB burdens. These data suggest that the progression to the cancer microenvironment is determined by genomic stress, providing clues for estimating cancer risk by reconsidering the fitness of cells in their microenvironment.
Keywords: Cancer; Mathematical biosciences; Molecular biology.
© 2024 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
References
LinkOut - more resources
Full Text Sources
Miscellaneous

