A negatively charged region within carboxy-terminal domain maintains proper CTCF DNA binding
- PMID: 39720519
- PMCID: PMC11667065
- DOI: 10.1016/j.isci.2024.111452
A negatively charged region within carboxy-terminal domain maintains proper CTCF DNA binding
Abstract
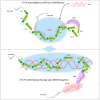
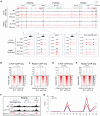
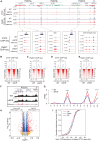
As an essential regulator of higher-order chromatin structures, CCCTC-binding factor (CTCF) is a highly conserved protein with a central DNA-binding domain of 11 tandem zinc fingers (ZFs), which are flanked by amino (N-) and carboxy (C-) terminal domains of intrinsically disordered regions. Here we report that CRISPR deletion of the entire C-terminal domain of alternating charge blocks decreases CTCF DNA binding but deletion of the C-terminal fragment of 116 amino acids results in increased CTCF DNA binding and aberrant gene regulation. Through a series of genetic targeting experiments, in conjunction with electrophoretic mobility shift assay (EMSA), circularized chromosome conformation capture (4C), qPCR, chromatin immunoprecipitation with sequencing (ChIP-seq), and assay for transposase-accessible chromatin with sequencing (ATAC-seq), we uncovered a negatively charged region (NCR) responsible for weakening CTCF DNA binding and chromatin accessibility. AlphaFold prediction suggests an autoinhibitory mechanism of CTCF via NCR as a flexible DNA mimic domain, possibly competing with DNA binding for the positively charged ZF surface area. Thus, the unstructured C-terminal domain plays an intricate role in maintaining proper CTCF-DNA interactions and 3D genome organization.
Keywords: Experimental systems for structural biology; Molecular Structure; Molecular physiology; Properties of biomolecules.
© 2024 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Neural network modeling of differential binding between wild-type and mutant CTCF reveals putative binding preferences for zinc fingers 1-2.BMC Genomics. 2022 Apr 12;23(1):295. doi: 10.1186/s12864-022-08486-9. BMC Genomics. 2022. PMID: 35410161 Free PMC article.
-
Dynamic Nature of CTCF Tandem 11 Zinc Fingers in Multivalent Recognition of DNA As Revealed by NMR Spectroscopy.J Phys Chem Lett. 2018 Jul 19;9(14):4020-4028. doi: 10.1021/acs.jpclett.8b01440. Epub 2018 Jul 6. J Phys Chem Lett. 2018. PMID: 29965776
-
7C: Computational Chromosome Conformation Capture by Correlation of ChIP-seq at CTCF motifs.BMC Genomics. 2019 Oct 25;20(1):777. doi: 10.1186/s12864-019-6088-0. BMC Genomics. 2019. PMID: 31653198 Free PMC article.
-
Discovering a binary CTCF code with a little help from BORIS.Nucleus. 2018 Jan 1;9(1):33-41. doi: 10.1080/19491034.2017.1394536. Epub 2017 Dec 5. Nucleus. 2018. PMID: 29077515 Free PMC article. Review.
-
CTCF shapes chromatin structure and gene expression in health and disease.EMBO Rep. 2022 Sep 5;23(9):e55146. doi: 10.15252/embr.202255146. Epub 2022 Aug 22. EMBO Rep. 2022. PMID: 35993175 Free PMC article. Review.
References
-
- Klenova E.M., Nicolas R.H., Paterson H.F., Carne A.F., Heath C.M., Goodwin G.H., Neiman P.E., Lobanenkov V.V. CTCF, a conserved nuclear factor required for optimal transcriptional activity of the chicken c-myc gene, is an 11-Zn-finger protein differentially expressed in multiple forms. Mol. Cell Biol. 1993;13:7612–7624. doi: 10.1128/mcb.13.12.7612-7624.1993. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

