Integrating machine learning, bioinformatics and experimental verification to identify a novel prognostic marker associated with tumor immune microenvironment in head and neck squamous carcinoma
- PMID: 39720726
- PMCID: PMC11666523
- DOI: 10.3389/fimmu.2024.1501486
Integrating machine learning, bioinformatics and experimental verification to identify a novel prognostic marker associated with tumor immune microenvironment in head and neck squamous carcinoma
Abstract
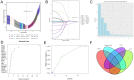
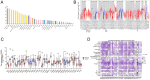
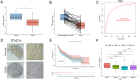
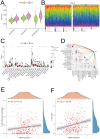
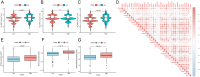
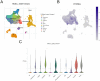
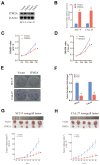
Head and neck squamous carcinoma (HNSC), characterized by a high degree of malignancy, develops in close association with the tumor immune microenvironment (TIME). Therefore, identifying effective targets related to HNSC and TIME is of paramount importance. Here, we employed the ESTIMATE algorithm to compute immune and stromal cell scores for HNSC samples from the TCGA database and identified differentially expressed genes (DEGs) based on these scores. Subsequently, we utilized four machine learning algorithms to identify four key genes: ITM2A, FOXP3, WIPF1, and RSPO1 from DEGs. Through a comprehensive pan-cancer analysis, our study identified aberrant expression of ITM2A across various tumor types, with a significant association with the TIME. Specifically, ITM2A expression was markedly reduced and correlated with poor prognosis in HNSC. Functional enrichment analysis revealed that ITM2A is implicated in multiple immune-related pathways, including immune-infiltrating cells, immune checkpoints, and immunotherapeutic responses. ITM2A expression was observed in various immune cell populations through single-cell analysis. Furthermore, we showed that ITM2A overexpression inhibited the growth of HNSC cells. Our results suggest that ITM2A may be a novel prognostic marker associated with TIME.
Keywords: HNSC; ITM2A; immune microenvironment; machine learning; prognostic.
Copyright © 2024 Zeng, Yang, Zhang, Li, Wang, Ma, Liao, Wang, Zeng and Zhang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Vos JL, Elbers J, Krijgsman O, Traets J, Qiao X, van der Leun AM, et al. Neoadjuvant immunotherapy with nivolumab and ipilimumab induces major pathological responses in patients with head and neck squamous cell carcinoma. Nat Commun. (2021) 12:7348. doi: 10.1038/s41467-021-26472-9 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

