Unraveling the impact of SARS-CoV-2 mutations on immunity: insights from innate immune recognition to antibody and T cell responses
- PMID: 39720734
- PMCID: PMC11666439
- DOI: 10.3389/fimmu.2024.1412873
Unraveling the impact of SARS-CoV-2 mutations on immunity: insights from innate immune recognition to antibody and T cell responses
Abstract
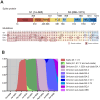
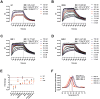
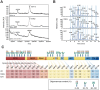
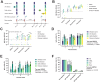
Throughout the COVID-19 pandemic, the emergence of new viral variants has challenged public health efforts, often evading antibody responses generated by infections and vaccinations. This immune escape has led to waves of breakthrough infections, raising questions about the efficacy and durability of immune protection. Here we focus on the impact of SARS-CoV-2 Delta and Omicron spike mutations on ACE-2 receptor binding, protein stability, and immune response evasion. Delta and Omicron variants had 3-5 times higher binding affinities to ACE-2 than the ancestral strain (KDwt = 23.4 nM, KDDelta = 8.08 nM, KDBA.1 = 4.77 nM, KDBA.2 = 4.47 nM). The pattern recognition molecule mannose-binding lectin (MBL) has been shown to recognize the spike protein. Here we found that MBL binding remained largely unchanged across the variants, even after introducing mutations at single glycan sites. Although MBL binding decreased post-vaccination, it increased by 2.6-fold upon IgG depletion, suggesting a compensatory or redundant role in immune recognition. Notably, we identified two glycan sites (N717 and N801) as potentially essential for the structural integrity of the spike protein. We also evaluated the antibody and T cell responses. Neutralization by serum immunoglobulins was predominantly mediated by IgG rather than IgA and was markedly impaired against the Delta (5.8-fold decrease) and Omicron variants BA.1 (17.4-fold) and BA.2 (14.2-fold). T cell responses, initially conserved, waned rapidly within 3 months post-Omicron infection. Our data suggests that immune imprinting may have hindered antibody and T cell responses toward the variants. Overall, despite decreased antibody neutralization, MBL recognition and T cell responses were generally unaffected by the variants. These findings extend our understanding of the complex interplay between viral adaptation and immune response, underscoring the importance of considering MBL interactions, immune imprinting, and viral evolution dynamics in developing new vaccine and treatment strategies.
Keywords: MBL; SARS-CoV-2; delta; immune imprinting; mannose-binding lectin; omicron; vaccine; variants of concern.
Copyright © 2024 Bayarri-Olmos, Sutta, Rosbjerg, Mortensen, Helgstrand, Nielsen, Pérez-Alós, González-García, Johnsen, Matthiesen, Egebjerg, Hansen, Sette, Grifoni, da Silva Antunes and Garred.
Conflict of interest statement
AlS is a consultant for AstraZeneca Pharmaceuticals, Calyptus Pharmaceuticals, Inc, Darwin Health, EmerVax, EUROIMMUN, F. Hoffman-La Roche Ltd, Fortress Biotech, Gilead Sciences, Granite bio., Gritstone Oncology, Guggenheim Securities, Moderna, Pfizer, RiverVest Venture Partners, and Turnstone Biologics. AG is a consultant for Pfizer. LJ has filed for patent protection for various aspects of T cell epitope and vaccine design work. MM, CH, PN, LJ, FM, TE are employed by Novo Nordisk A/S. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- WHO . Tracking SARS-CoV-2 variants (2023). Available online at: https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/ (Accessed 10 August 2022).
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

