Emergence of Omicron FN.1 a descendent of BQ.1.1 in Botswana
- PMID: 39720788
- PMCID: PMC11666700
- DOI: 10.1093/ve/veae095
Emergence of Omicron FN.1 a descendent of BQ.1.1 in Botswana
Abstract
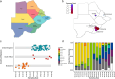
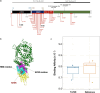
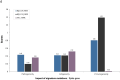
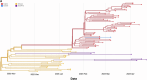
Botswana, like the rest of the world, has been significantly impacted by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). In December 2022, we detected a monophyletic cluster of genomes comprising a sublineage of the Omicron variant of concern (VOC) designated as B.1.1.529.5.3.1.1.1.1.1.1.74.1 (alias FN.1, clade 22E). These genomes were sourced from both epidemiologically linked and unlinked samples collected in three close locations within the district of Greater Gaborone. In this study, we assessed the worldwide prevalence of the FN.1 lineage, evaluated its mutational profile, and conducted a phylogeographic analysis to reveal its global dispersal dynamics. Among approximately 16 million publicly available SARS-CoV-2 sequences generated by 30 September 2023, only 87 were of the FN.1 lineage, including 22 from Botswana, 6 from South Africa, and 59 from the UK. The estimated time to the most recent common ancestor of the 87 FN.1 sequences was 22 October 2022 [95% highest posterior density: 2 September 2022-24 November 2022], with the earliest of the 22 Botswana sequences having been sampled on 7 December 2022. Discrete trait reconstruction of FN.1 identified Botswana as the most probable place of origin. The FN.1 lineage is derived from the BQ.1.1 lineage and carries two missense variants in the spike protein, S:K182E in NTD and S:T478R in RDB. Among the over 90 SARS-CoV-2 lineages circulating in Botswana between September 2020 and July 2023, FN.1 was most closely related to BQ.1.1.74 based on maximum likelihood phylogenetic inference, differing only by the S:K182E mutation found in FN.1. Given the early detection of numerous novel variants from Botswana and its neighbouring countries, our study underscores the necessity of continuous surveillance to monitor the emergence of potential VOCs, integrating molecular and spatial data to identify dissemination patterns enhancing preparedness efforts.
Keywords: Africa; Botswana; Omicron FN.1; SARS-CoV-2; phylodynamics.
© The Author(s) 2024. Published by Oxford University Press.
Conflict of interest statement
None declared.
Figures

Similar articles
-
Application of a high-resolution melt assay for monitoring SARS-CoV-2 variants in Burkina Faso and Kenya.mSphere. 2025 Jun 25;10(6):e0002725. doi: 10.1128/msphere.00027-25. Epub 2025 May 29. mSphere. 2025. PMID: 40439429 Free PMC article.
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
-
Emergence and spread of the SARS-CoV-2 omicron (BA.1) variant across Africa: an observational study.Lancet Glob Health. 2025 Feb;13(2):e256-e267. doi: 10.1016/S2214-109X(24)00419-4. Lancet Glob Health. 2025. PMID: 39890226
-
Antibody tests for identification of current and past infection with SARS-CoV-2.Cochrane Database Syst Rev. 2022 Nov 17;11(11):CD013652. doi: 10.1002/14651858.CD013652.pub2. Cochrane Database Syst Rev. 2022. PMID: 36394900 Free PMC article.
-
Genomic epidemiology of SARS-CoV-2 in Norfolk, UK, March 2020-December 2022.Microb Genom. 2025 Jul;11(7):001435. doi: 10.1099/mgen.0.001435. Microb Genom. 2025. PMID: 40663468 Free PMC article.
References
-
- Aksamentov I, Roemer C, Hodcroft E. et al. Nextclade: clade assignment, mutation calling and quality control for viral genomes. J Open Source Softw 2021;6:3773. doi: 10.21105/joss.03773 - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

